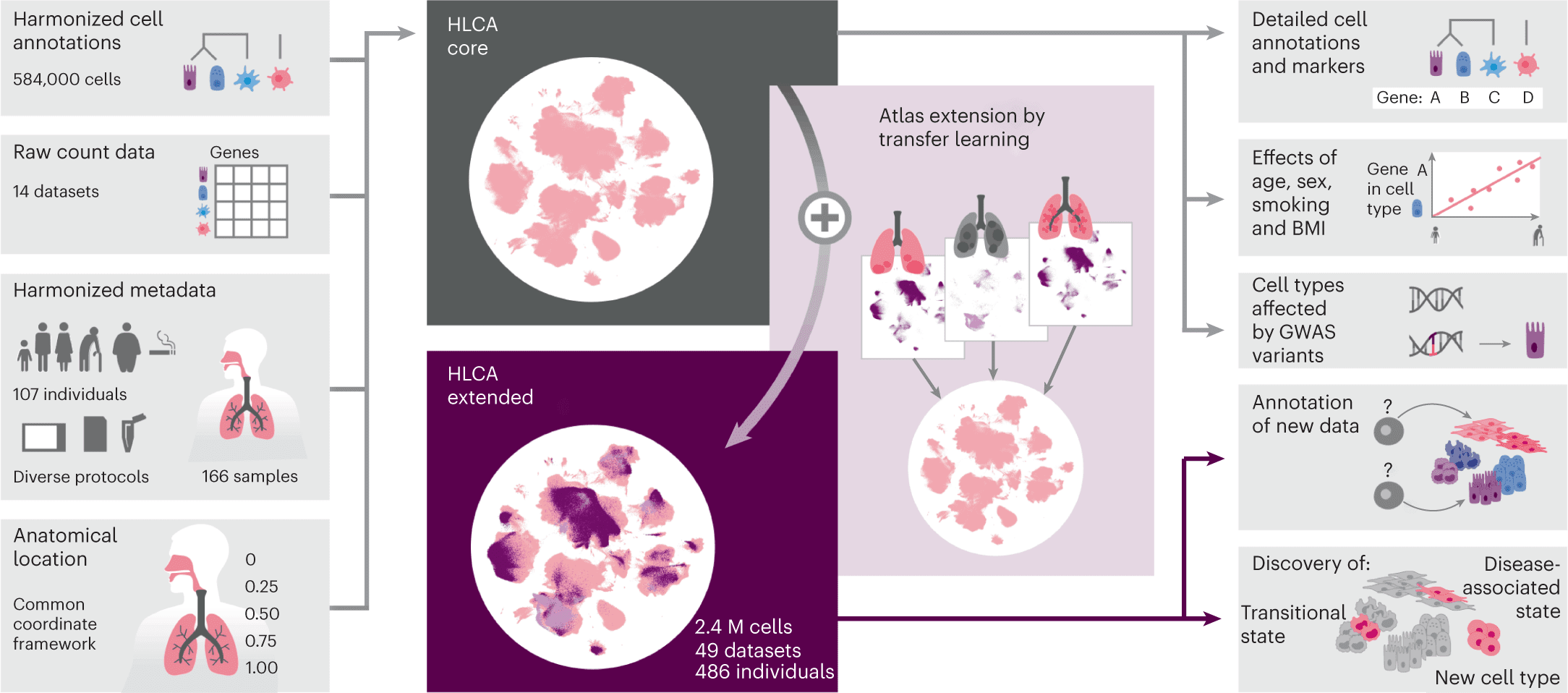
The Human Lung Cell Atlas (HLCA) represents comprehensive single-cell transcriptomics of the human respiratory system, which aims to resolve the limitations in previous atlases. By integrating data from 486 donors and 49 datasets, encompassing 2.4 million cells and donor metadata, the HLCA seeks to capture individualistic variations, overcome limited samples, and mitigate technical biases. Its purpose is to provide a deeper understanding of health and diseases by comprehensively representing the population’s variation. Additionally, the HLCA serves as an unbiased, accurate, and extensive reference for future studies and applications in the field. The Human Lung Cell Atlas (HLCA), a vital new tool for lung researchers, reveals the enormous variety of cell types present in the lung as well as significant distinctions between health and sickness.
Insights into Different Cell Types
The combined diverse sets of data were re-annotated to serve as a curated reference through refinement of earlier understandings, revamping characteristics of cell types, and universal marker genes of healthy lung tissue.
The group of researchers detected six types of cells in human lungs reported rarely in former studies such as dendritic cells, hematopoietic stem cells, highly proliferative hillock-like epithelial cells, and alveolar type 0 cells preterminal bronchiole secretory cells. Among them, hillock-like epithelial cells were detected for the first time in adult lungs and, thus, improved the designation of rare and unknown cell types. Moreover, the proportion of cell types in different parts of the respiratory system was evaluated, such as ionocytes were present at comparable proportions in the airway epithelium and largely absent in lung parenchyma.
Characterization of Gene Expression Attributed by Variable Factors
Factors such as age, gender, smoking history, BMI, and ethnicity influence cellular transcriptomics. The data analysis approached to generate a generalistic understanding of the featurization about its impacts at the molecular level. Other aspects such as cell types, anatomical location of the cell, technical covariates caused by mitochondrial genes, and variation in experimental protocols were considered to infer contributions of the variable factors in the expression profile.
For instance, smoking status was found to be affecting transcriptome profiles of innate lymphoid cells, BMI is associated with fluctuations of expression level in B and T cells; downregulation of oxidative phosphorylation, interleukin1 signal, etc. was noted in distal cells. Higher BMI was correlated with the downregulation of the insulin response pathway in secretory cells and insulin resistance observed in donors with obesity. Higher BMI is linked with perturbations in different gene expressions in plasma cells, consistently indicating obesity as a risk factor for plasma cell malignancy.
Identification of Disease Condition
Discerning disease-specific conditions and alternations can be achieved through HCLA. Mapping scRNA-seq (Single-Cell RNA Sequencing) data from lung cancer samples against HCLA core data assisted in revealing erroneous notions about certain markers, such as ACKR1 and EDNRB. HCLA can serve as a reference for Genome-wide Association Studies (GWASs) to designate disease-related nucleotide variants. For example, researchers identified a significant association of lung T cells with asthma-associated single-nucleotide polymorphisms (SNPs). Comprising diverse datasets, HLSA can provide accurate contextualization of different diseases. Automated mapping of new data to the HLCA core can be done by any user via an interactive web portal of HLCA or using code tutorials.
The HLCA contains data across more than ten lung diseases, enabling feasibility to uncover cellular states specific to disease conditions. The researchers have provided cell-type signature matrices. Therefore, HLCA can also assist in recognizing cell type and its expression profile affected particularly in certain diseases. For example, the researchers demonstrated common genetic markers in Idiopathic pulmonary fibrosis (IPF) specific alveolar fibroblast state. Disease-specific variability of cell type proportions can be deciphered from the HLCA resources. For example, smooth muscle displayed the largest shift in proportion in patients with severe COPD (Chronic Obstructive Pulmonary Disorder). Disease subtypes and characteristic patterns can be demarcated utilizing HLCA.
Conclusion
HLCA is a collaborative approach to building the largest and most comprehensive cell map of the human lung. Owing to the unprecedented range of diversity in datasets, HLCA will be able to fulfill the crucial need for ‘reference’ absolutely essential for speeding up further studies. The combination of corresponding anatomical, physiological, and donor covariates will offer the path of new research prospects. HLCA core can serve as a healthy control and contextualize disease data by mapping disease-specific alterations. Linking genetic signatures with cell type and cell state will provide the foundation for understanding disease progression. HCLA was prepared using the top-performing integration method, scANVI, to achieve high-quality integrated data. So it can be utilized to annotate novel data sets and identify new cell types. Most importantly, HCLA will help to understand the molecular targets for designing effective drugs and drug repurposing.
Indeed, the HLCA is a live resource that will be continuously updated and refined. HLCA can serve as a community- and data-driven platform for open discussion on lung cell identities. The scientists anticipate that the HLCA will be completed in two phases: when no new cell types can be identified and when all individual variations across all populations will be fully represented. Although HLCA possesses an intrinsic limitation that 65% of the datasets belong to European ethnicity, as researchers mentioned, this unintended bias will also be repaired in the near future. Nonetheless, HCLA is a cornerstone for unlocking new aspects of respiratory health, and function, a promising resource for investigating COPD, lung carcinoma, pulmonary fibrosis, etc., diseases and finding new approaches for drug development.
Article Source: Reference Paper | Access HCLA at GitHub
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.