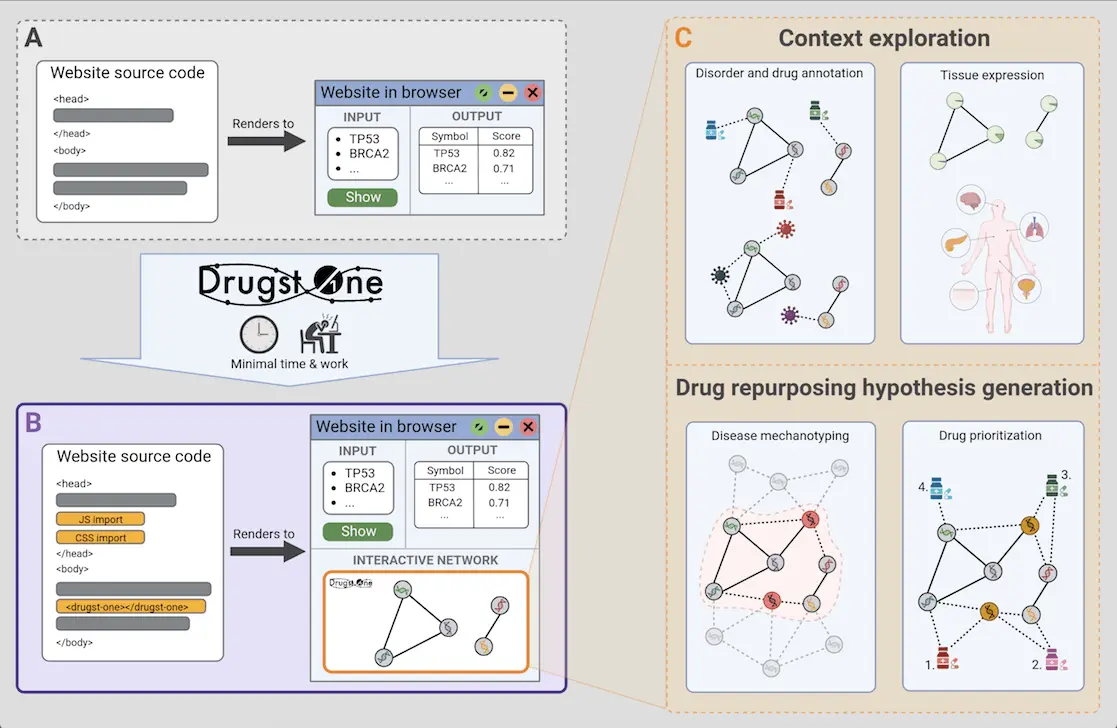
Researchers from the University of Hamburg, Germany, in collaboration with researchers from Denmark, Canada, the USA, Spain, Slovakia, and Israel, have designed Drugst.One, a platform that transforms complex computational tools into user-friendly, web-based utilities for finding novel applications for existing drugs. With as little as three lines of code, Drugst.One can convert any gene or protein-based computational tool into a powerful, web-based toolkit that helps in visualizing how medicines interact with proteins and diseases, which can aid in repurposing already existing drugs. It has already been implemented with 21 different tools, including ones specializing in drug repurposing for irritable bowel disease (IBD) and investigating the smooth muscle cell (SMC) proliferation pathway.
A Brief Background of Visualization of Protein-Drug Interaction
Due to increasing medical demands, technology, over the years, has advanced and catapulted the emergence of tools that enable the identification of the genes or proteins responsible for a particular disease along with the visualization of their interactions contributing to the progression of the disease. This visualization enhances the precision of pharmaceutical treatments and minimizes side effects. It also enables drug repurposing, which eliminates the expenses required for developing new drugs.
However, developing such tools is not devoid of difficulties and drawbacks including:
- Combining credible biomedical data from various sources
- Developing flexible graphical user interfaces for data analysis and visualizations.
- Existing software such as Cytoscape cannot visualize biological networks without local installation for each user.
- Online alternatives dependent on browser compatibility are inconvenient for researchers lacking sufficient web-development expertise.
- The acquisition and harmonization of diverse biomedical data is a time-consuming task.
- The databases storing biomedical data require frequent maintenance and regular updates.
- If exploring the biological networks is not considered additional work, then new tools are being created from scratch, which can lead to variations in the quality of the results produced by the individual tools.
A One-Stop Solution to All the Challenges: Drugst.One
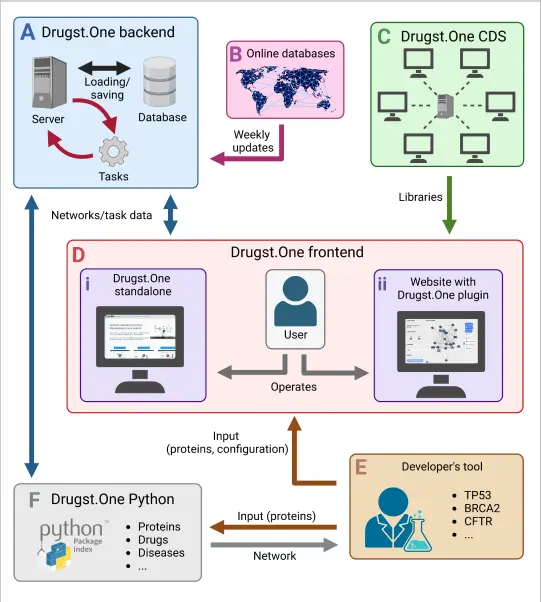
Image Source: https://arxiv.org/abs/2305.15453
To circumvent the challenges mentioned above, researcher Jan Baumbach and his team have developed Drugst.One, which reduces the technical complexities like maintaining and debugging the computational tools based on gene and protein data and simplifies the visualization of biological interactions. Requiring minimal coding, Drugst.One can be added to any gene or protein-based computational biology tool, converting it into a robust online toolkit for exploratory data analysis, visualization, and drug-repurposing candidate identification.
Drugst.One bridges the gap between exploring disease mechanisms and creating hypotheses for putting existing drugs into new uses. It incorporates various state-of-the-art databases and undergoes weekly updates ensuring the relevance of its database for continuously changing data. As of now, it has been integrated into 21 biomedical software such as mirDIP and WikiPathways. It takes a list of proteins or genes in HGNC (HUGO Gene Nomenclature Committee), UniProt, Ensembl, or Entrez ID format and combines them with other relevant data to generate a map of how they interact with each other and with various drugs.
The Origin, Mechanism, and Features of Drugst.One
The origins of Drugst.One can be traced to CoVex and CADDIE, which are network-based drug repurposing tools. CoVex is used for SARS-CoV-2, and CADDIE is used for cancer. These tools have the same underlying algorithms, which have been tested and proven effective. These algorithms include identification algorithms and ranking algorithms.
Drugst.One works in the following manner:-
- Input: A list of genes or proteins related to a disease is fed as the input.
- Processing: Drugst.One combines these genes or proteins with other relevant data to comprehend how they work together and their interaction with drugs.
- Annotating: Drugst.One labels these proteins or genes with crucial information, like which drugs target them or how they contribute to the disease.
- Visualisation: Drugst.One provides a visual representation through a network among the proteins/genes and between the proteins/genes and drugs, highlighting which drugs might work for the disease.
- Target Identification: Drugst.One makes use of the identification algorithms and identifies the target proteins/genes which are related to the disease from the network.
- Drug Repurposing: It also employs a ranking algorithm to rank which drugs can potentially be repurposed.
There are many features of Drugst.One which takes exploratory data analysis of proteins/genes and drugs up a notch. Such features include:
- It allows the exploration of the tissues or organs which demonstrate higher expression of the proteins/genes related to the disease.
- It provides multiple sources of data.
- It provides interactive mode, reset mode as well as export options for the network.
- It also helps in checking whether certain biological functions or pathways are represented in a set of genes or proteins.
- It allows searching for well-known pathways involving the same proteins.
- It enables user involvement by allowing them to add their own knowledge through the manipulation of input.
- It also allows the user to implement identification and ranking algorithms of their choice.
A Glimpse at the Vast Applicability of Drugst.One
Drugst.One has proven its adaptability and resourcefulness with multiple computational tools and across various aspects of drug development. Some of its applications include:
Drugst.One with ROBUST-Web: The computational tool ROBUST-Web helps researchers study diseases using a special technique to find crucial regions in the network of genes related to the disease. After that, those parts are sent to Drugst.One which provides a visual representation aiding in gaining clarity and comprehension for researchers. It also annotates and suggests possible treatments.
Drugst.One with BiCoN: BiCoN is a computational tool for patient stratification and disease mechanism identification. BiCoN takes molecular networks as input and splits a group of patients into sub-groups based on certain characteristics. It also identifies a subnetwork of molecules (genes) showing differential activity in subgroups of patients, which can serve as biomarkers or potential drug targets. The subnetwork is sent to Drugst.One which interactively presents the results, enabling the exploration of possible treatments and drug options.
Drugst.One with WikiPathways: WikiPathways is used by scientists to share information about how genes work together in pathways that may play a role in diseases and treatments. This information is sent to Drugst.One which demonstrates how those genes interact with drugs and also allows further exploration of the same.
Drugst.One for deciphering disease mechanisms: Visualizing the SMC pathway on Drugst.One interface, researchers could better comprehend the mechanisms of cardiovascular disease (CVDs). In the visual representation, several CVD-related nodes appear, such as cardiomyopathy, coronary artery disease, etc. SMCs are vital for proper vascular functioning and play a role in atherosclerosis, hypertension, and heart attacks. Using data from the Genotype-Tissue Expression (GTEx) project, Drugst.One shows that SMC genes have higher expression in tissues responsible for physical movement like the heart, lungs, etc.
Drugst.One for Drug Repurposing: Existing drugs for the CVDs have not been identified yet. However, some anti-cancer drugs, like sunitinib, erlotinib, midostaurin, and ruxolitinib, which target the calcium/calmodulin-dependent protein kinase II delta (CAMK2D) protein, have been found. Interestingly, CAMK2D is involved in calcium signaling, which maintains SMC function. With Drugst.One’s algorithms, the drug nintedanib, has been found to be indirectly connected to the SMC pathway and has shown promise in affecting pulmonary arterial SMC and intestinal SMC.
Conclusion
Researchers continuously strive to find new drugs and treatments for illnesses, but finding and developing new therapeutics is costly as well as cumbersome. In such cases, Drugst.One comes to the rescue. It works with various websites and tools that researchers already use, converts complicated biomedical data into clear, interactive visual representations, and speeds up the process of finding new therapeutics. It also helps in gaining higher clarity in disease mechanisms and repurposing already existing drugs, saving money and time. However, it should be remembered that the predictions of Drugst.One are just ideas that need to be experimentally validated by real experts. Nevertheless, Drugst.One gives researchers a much-needed direction to take in drug development, elevating drug discovery to even greater heights.
Article Source: Reference Paper | Drugst.One: Website
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Neegar is a consulting scientific content writing intern at CBIRT. She's a final-year student pursuing a B.Tech in Biotechnology at Odisha University of Technology and Research. Neegar's enthusiasm is sparked by the dynamic and interdisciplinary aspects of bioinformatics. She possesses a remarkable ability to elucidate intricate concepts using accessible language. Consequently, she aspires to amalgamate her proficiency in bioinformatics with her passion for writing, aiming to convey pioneering breakthroughs and innovations in the field of bioinformatics in a comprehensible manner to a wide audience.