Scientists from the University of Helsinki, Finland, developed a drug repurposing scoring algorithm, DrugRepo, that predicted 6229 new drug-disease relationships across 545 diseases for 516 approved drugs. The effectiveness of the algorithm was validated through evaluation studies that identified ≥0.4 as an appropriate threshold for discovering new candidate drugs. The study is also accompanied by a DrugRepo GUI that allows users to download repurposable drug candidates for a given disease.
One of the earliest and the most popular cases of drug repurposing would be that of sildenafil, originally marketed as a hypertensive drug. It was turned into the remedy for what was once thought of as its side effect, penile erections, becoming Viagra used to treat erectile dysfunction. Another story is of thalidomide, a sedative for morning sickness that was withdrawn from the market in association with severe birth defects. Later studies lead to it being reintroduced into the market as an anticancer drug that targets angiogenesis.
One of the earliest and the most popular cases of drug repurposing would be that of sildenafil, originally marketed as a hypertensive drug. It was turned into a remedy for what was once thought of as its side effect, penile erections, becoming Viagra used to treat erectile dysfunction. Another story is of thalidomide, a sedative for morning sickness that was withdrawn from the market in association with severe congenital disabilities. Later studies led to it being reintroduced into the market as an anticancer drug that targets angiogenesis.
With the ever-increasing horde of disease- and drug-related data and computerized tools to analyze them, drug discovery has attained a new cape of computer-aided drug design. New purposes for already existing drug candidates(drug repurposing) are no more a matter of serendipity but a systemic analysis through the sea of data repositories to design computational methods to study drug interactions with newer targets. The most recent study in this field originates from the University of Helsinki, Finland, where 516 drugs were repurposed, predicting 6229 possible drug-disease pairs through a novel scoring algorithm, DrugRepo.
The Making of DrugRepo
The DrugRepo scoring algorithm is an aggregate of three highly specific scoring functions:
- Overlapping Compound-Target Score (OCTS): It scores each compound based on the overlap between the drug-target interaction profiles of the said compound and a drug approved for the given disease. The more targets common between the two, the higher the OCTS score.
- Tanimoto Coefficient (TC): It assesses the structural similarity between the approved drug and the investigational compound through their ECFP4 fingerprints.
- Compound-Disease Score (CDS): It studies the disease-compound association through disease-gene and protein-protein interaction networks. The disease-gene network identifies the relevant genes and hence the target proteins associated with a disease. Protein-protein interaction calculates the average distance between the candidate compound and the target protein.
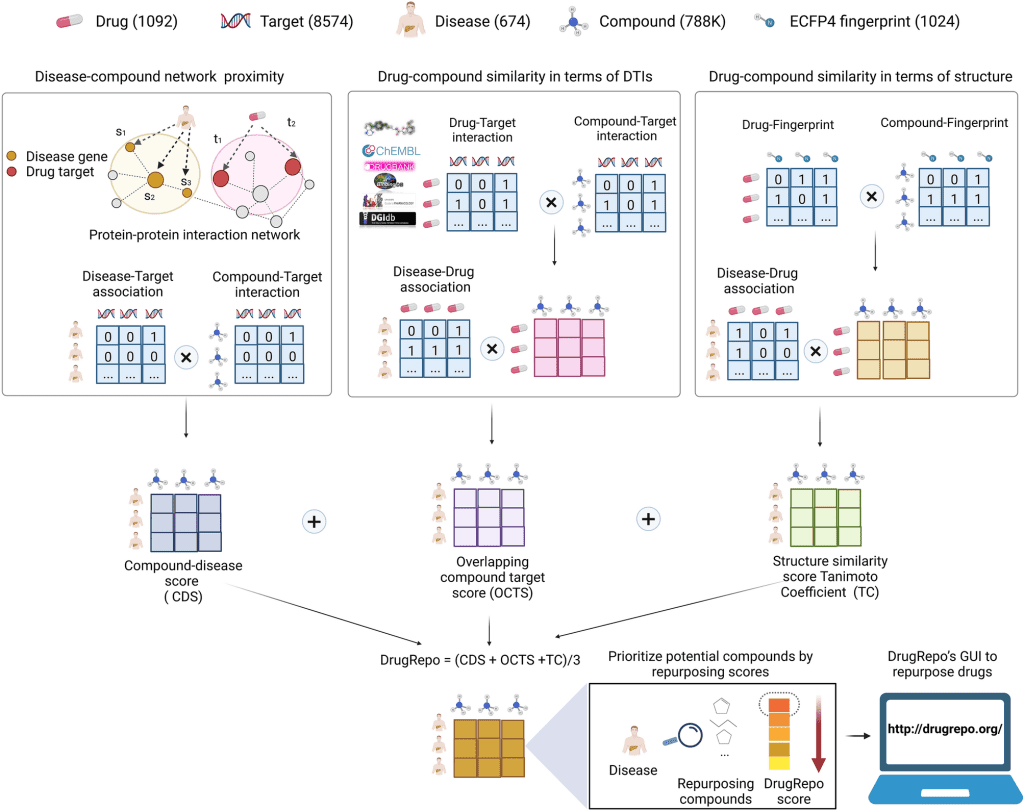
Image source: https://doi.org/10.1038/s41598-022-24980-2
The first two scoring functions approximate the disease-compound association through the compound-drug association. The third assesses this directly through the compound associated with the target proteins of the disease. The three scores are normalized to a value between 0 and 1. The scores from the three functions are averaged to obtain a DrugRepo score for each compound for a particular disease.
A total of 788,078 investigational compounds were pooled from five different databases after filtering out overlapping compounds. Around 1091 drugs approved for the treatment of 3757 drug indications linked to 669 diseases were obtained from the clinical trials database. For each compound, the DrugRepo score was calculated by comparing it to the 1091 approved drugs. In a similar fashion, DrugRepo scores were also computed for a drug approved for a particular disease by comparing it with drugs approved for another disease. The results of this study, along with DrugRepo scores of the repurposed compounds for each disease, are reported through a user-friendly web tool whose link is given below this article.
Evaluation of DrugRepo
The translational impact of the scoring algorithm was assessed by studying the validity of the repurposed compounds. This was performed by two methods:
- Cross-referencing with disease-compound associations in Comparative Toxicogenomic Database (CTD): CTD contains compound-disease associations for 3941 compounds and 6119 diseases by inferring disease-gene and drug-target relationships. The datasets and scoring methods used for CTD are different from that of the current study. Leaving out diseases that cannot be matched to common identifiers reduced CTD disease count to 605, out of which 118 diseases had more than one repurposed compound in common with DrugRepo. This suggests DrugRepo’s effectiveness in repurposing compounds for several diseases.
- Matching repurposed compounds with compounds under clinical trials: It was performed to observe whether DrugRepo could enrich compounds in parallel with experimental studies. Across nine cancers, DrugRepo identified 186 compounds that have completed phase I or phase II trials. The study also established a DrugRepo score ≥ 0.4 as the appropriate threshold for detecting possible drug candidates.
With the DrugRepo score ≥0.4, the study identified potential drug candidates both among investigational compounds and approved drugs for the diseases under study. Five hundred and sixteen drugs have been repurposed for 545 diseases, where a few have a DrugRepo score ≥0.9. Some of these are under different phases of clinical trials, while some have not even been considered for the repurposed disease. The DrugRepo GUI divulges this information as a means to encourage new experimental studies addressing these gaps.
Limitations of DrugRepo
The greatest setback the authors faced in the study was the lack of drug target profile data for computing the OCTS score. The average number of targets for approved drugs was seven, even after cumulating the data from five of the most extensive databases. Yet, this is believed to improve with the new releases of additional drug-target information in these databases, which could improve the repurposing potential of the algorithm. Furthermore, the authors are already planning an improved version of DrugoRepo by including additional scoring functions, with gene expression data being one of the candidates.
Article Source: Reference Paper | DrugRepo GUI
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Catherene Tomy is a consulting Content Writing Intern at the Centre of Bioinformatics Research and Technology (CBIRT). She has a master’s degree in Molecular Medicine from Amrita University with research experience in the fields of bioinformatics, cell biology, and molecular biology. She loves to pull apart complex concepts and weave a story around them.
.







