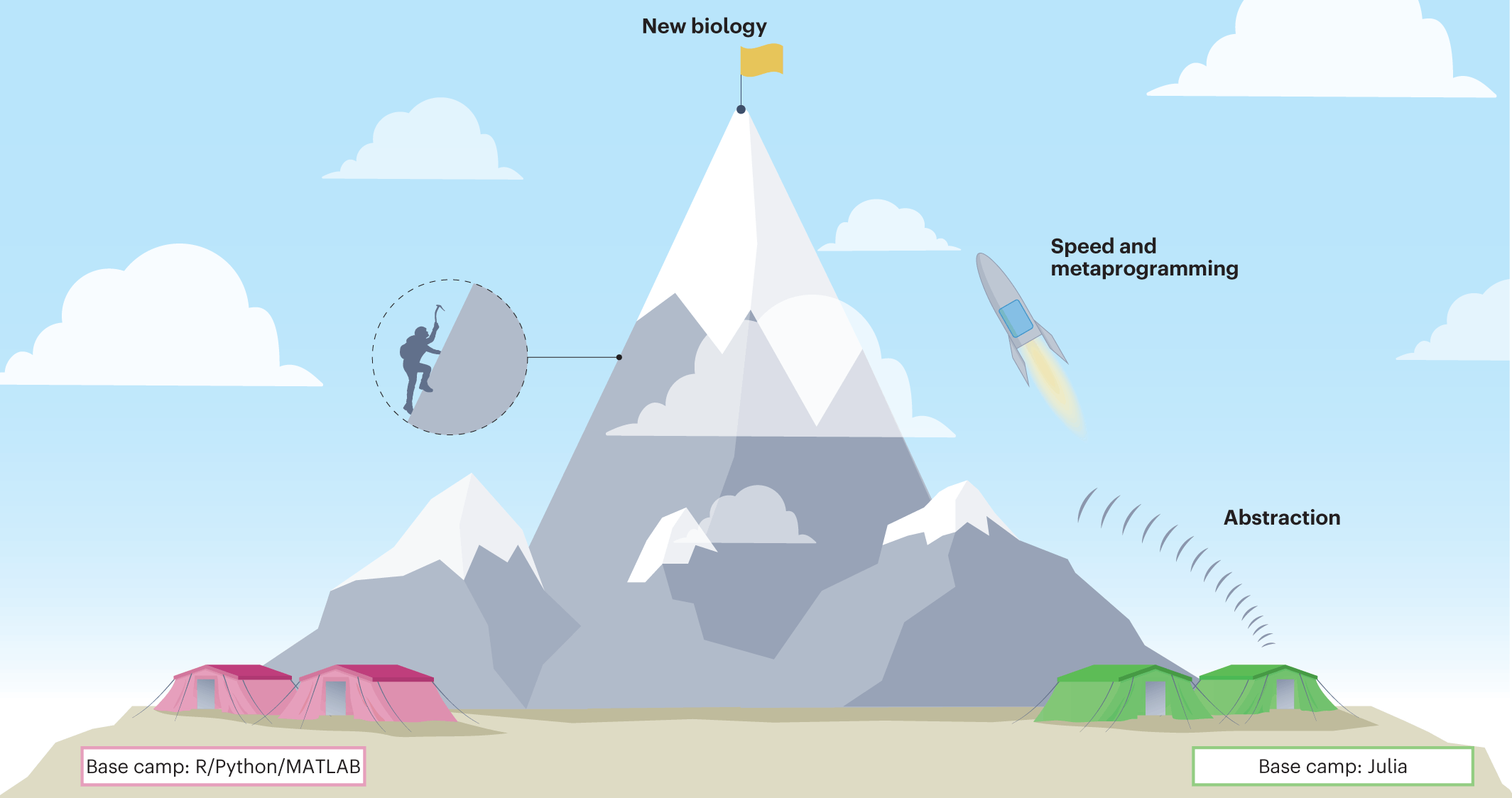
As researchers gather increasingly complex and extensive data, it is also becoming increasingly difficult to accurately and efficiently process and analyze everything. Julia, a relatively new programming language, presents unique benefits and features, making it an ideal solution for this challenge. Its three key features make it stand out from other existing languages, i.e., speed, abstraction, and metaprogramming abilities. Julia has great potential and can perform computational tasks in systems biology in a way that was not possible previously!
The Importance of Computational Tools in Biosciences and its Challenges
Computers are tools that allow us to perform tasks more efficiently and provide crucial insights into biological systems and data. They have become increasingly essential in solving biological problems and have given rise to the computational biology and bioinformatics fields. Without computers, many significant projects and scientific breakthroughs, such as the 1000 Genomes Project and vaccine development, would have been rather impossible.
In addition, programming languages are tools that allow us to command computers, instructing them what to do. Specific languages are meant for specific tasks. In biomedical research, R and Python are the predominant languages; however, computationally demanding research often depends on Fortran or C/C++. In practice, such computationally demanding/intensive research is initially prototyped in R, Python, or MATLAB and eventually translated into the faster languages C/C++ or Fortran. This is called the two-language approach.
This approach has been successful but does have its limitations. This is because directly translating the code may not always be ideal, and also, using the features of C/C++ or Fortran may require a total rewrite of the algorithm to validate faster implementation or better scaling. Therefore, the two-language approach would necessitate expertise and skills across both languages and thorough code testing in both languages.
Julia Programming Language
Recently, a paper published in Nature Methods discussed the features and relevance of a relatively new programming language, Julia, to address the existing issues in computational biosciences highlighted by the two language problems.
Julia has been created and designed to be an easy-to-program and quick-to-implement programming language. It overcomes the two-language problem because users do not have to balance or choose between the ease of use and high performance. Biological systems and data, given their complexity, need flexible programming languages that can be used to describe and model them mathematically. Julia possesses three key features—speed, abstraction, and metaprogramming—which make the language ideal for meeting the demands of biomedical science. Julia’s speed allows faster computation, and its abstraction and metaprogramming abilities enable the connection of highly structured data types, making it a crucial tool for researchers in relevant fields.
The Three Key Features of Julia
- Speed
In computational practices and programming languages, speed is not merely crucial for completing analyses quickly but is instead vital to analyze large and extensive datasets, which are instead becoming the current norm in the field of biomedical research. In fact, slow computations act as a limiting factor when used to analyze large datasets repeatedly. Additionally, with slow implementation, it would be impossible to stimulate large and complex computational models. Hence, fast computation is very important and can be highlighted by the fact that digital twins in precision medicine are useless without fast computation.
Julia can offer the fast speed of statistically compiled languages like C/C++ and Fortran but with additional higher-level language characteristics. Julia exploits fast development with rapid run-time performance, making it suitable for algorithm/method designing and computationally intensive uses.
To demonstrate the fascinating speed of Julia, the paper discusses an example of using Julia’s speed to address the statistically demanding task of network interference from single-cell data. In particular, Julia’s ‘InformationMeasures.jl’ package implementation of the mutual information measure was compared with the R package implementation. It was seen that with datasets containing 3500 genes and 600 cells, R requires 2.5 hours as opposed to Julia’s remarkable 134 seconds. In real-world applications, up to 400-fold speed differences could be observed between R and Julia.
- Abstraction
Julia demonstrates a high level of abstraction, which benefits its abilities in software development. The authors use the example of pipettors to highlight how Julia interfaces can minimize efforts in adapting to new data types and switching between different implementations of the same interface. Since biological data are complex and heterogeneous, it represents a significant challenge for software developers and data analysis pipelines. However, Julia’s abstraction abilities allow specialization and generalization through characteristics such as abstract interfaces and generic functions that can utilize unique data formats with differing intrinsic features without a performance penalty.
The paper uses an example of a structural bioinformatics pipeline to demonstrate Julia’s abstraction abilities. It was seen that Julia’s composability feature allows users to customize packages to meet the specific needs of their research. Hence, Julia’s abstraction allows for improvements in any of these packages, which would benefit users of other packages. Julia’s abstraction abilities enable interoperability and longevity of code, something not implemented by languages like Python, R and C/C++.
- Metaprogramming
Julia also demonstrates remarkable metaprogramming abilities/techniques, i.e., the techniques allowing a program to modify its own source code during run time. Julia’s “hygenic macros” feature is what allows for this technique. The macros code templates can be manipulated during runtime, allowing repetitive code to be generated effectively and efficiently. These metaprogramming abilities are crucial in modeling biological systems since they enable new approaches to formulating and adapting mathematical models of the systems. In fact, Julia’s ‘Catalyst.jl’ package uses a single block of code to generate mathematical models specified in terms of biochemical reactions with the associated rates. Additionally, the models can be analyzed using several frameworks, for instance, ordinary or stochastic differential equations. Finally, Julia’s package ecosystem allows the fitting of models to data or even supports the estimation of parameters.
Conclusion
In conclusion, Julia is a relatively new and modern language that is fast and flexible, with a state-of-the-art package manager. Given its growing number of users and advantages in specific domains, Julia is expected to be the next chapter in quantitative and computational life sciences! However, the authors acknowledge that convenience and user community are essential factors where R, Python, and C/C++ clearly gain an advantage and acknowledge the need for even more Julia users to facilitate an overall switch to this language.
Article Source: Reference Paper
Learn More:
Diyan Jain is a second-year undergraduate majoring in Biotechnology at Imperial College, London, and currently interning as a scientific content writer at CBIRT. His passion for writing and science has led him to pursue this opportunity to communicate cutting-edge research and discoveries engagingly to a broader public. Diyan is also working on a personal research project to evaluate the potential for genome sequencing studies and GWAS to identify disease likelihood and determine personalized treatments. With his fascination for bioinformatics and science communication, he is committed to delivering high-quality content a CBIRT.