Scientists from Columbia University, New York, have reported the possibility of automated cell type annotation in single-cell RNA seq (scRNA-seq) analysis using GPT-4, a chatbot. While cell type annotation is a significant step in the pipeline of sc-RNAseq analysis, it is often time-consuming and requires manual annotation using canonical marker genes. Automated cell-type annotation typically involves additional computational pipelines and the acquisition of high-quality reference datasets. The authors demonstrate the usage of GPT-4, a highly advanced and multimodal language model, in annotating cell types automatically using marker gene information from the standard scRNA-seq pipelines. Annotations generated by GPT-4 exhibit strong agreement with manual annotations when applied to hundreds of different cell and tissue types. The cell-type annotation will no longer be time-consuming, as exhibited by GPT-4’s fast generation of cell-type annotations.
Cell-type Annotation in scRNA-seq Analysis: Manual vs. Automated
Cell-type annotation is a crucial step in any scRNA-seq pipeline. Cell-type annotation determines the heterogeneity within the cell population and provides an understanding of the diverse functions of the different cell populations within complex tissues. Seurat and Scanpy, softwares for single-cell analysis, routinely employ manual cell-type annotation. These tools typically assign single cells to clusters, which are obtained using cell clustering techniques. Studies about identifying differentially expressed genes across cell clusters are also conducted. Comparison by a human expert of canonical cell type markers with differential gene expression information leads to successful cell type annotation to each cell cluster. The process is cumbersome owing to the prior knowledge of canonical cell type markers as well as being time-consuming.
Several tools have been developed over the years for automated cell types annotation, such as SingleCellNet, LAmbDA, and scmap. However, manual cell-type annotation using marker gene information is the most widely used option for any single-cell analysis pipeline.
GPT: Generative Pre-Trained Transformers
GPTs, Generative Pre-trained Transformers, are large language models trained on massive amounts of data to produce human-like text given the context. GPTs such as GPT-3, ChatGPT, and GPT-4 are among the most advanced language models, and recent studies have demonstrated the competitive performance of GPTs in answering biomedical questions.
GPT-4 has been shown to surpass ChatGPT’s advanced reasoning capabilities. GPT-4 has proven to solve difficult problems with greater accuracy owing to its broader general knowledge and problem-solving capabilities. Several organizations have collaborated with OpenAI’s GPT-4, which includes Duolingo, Morgan Stanley, Khan Academy, and even the Government of Iceland, for its language preservation, as is mentioned on the GPT-4 homepage. Such is the broad span of usability of GPT-4, and hence it is evident that scientists and researchers would try and find its usage in scientific research and work.
Can we use GPT-4 for Cell-type Annotation?
The authors hypothesize that GPT-4, one of the most advanced language models, can be applied to the task of annotating single cell types using marker gene information. While other automated cell-type annotation pipelines require collecting high-quality reference datasets as well as developing additional computational pipelines, GPT-4 seamlessly integrates into existing scRNA-seq analysis pipelines such as Scanpy and Seurat. It is also cost-effective and offers the option of fine-tuning the generated annotations using human expertise. The vast landscape of data that GPT-4 is trained on enables it to be applied to various cell types and tissue types, while other automated pipelines are restricted by the usage of specific reference datasets. Moreover, the chatbot’s characteristics are highly user-friendly and enable incorporating user feedback for iterative answer improvement.
How Exactly do we Chat and Annotate Cell-types?
The author set out to test their hypothesis regarding using GPT-4 for annotating cell types. The systematic analysis includes the assessment of GPT -4’s performance on cell-type annotation tasks for five datasets, including hundreds of cell types and tissue types across both human and mouse lineages. The inputs for GPT-4 included canonical marker genes as identified in the dataset for this article, as well as computationally identified genes for the four sc-RNAseq datasets, viz., Azimuth by HuBMAP, Human Cell Atlas (HCA), Human Cell Landscape (HCL) and Mouse Cell Atlas (MCA).
Cell-type annotation was performed for the HCL and MCA datasets by aggregating all tissues, whereas for the other datasets, the annotation task was performed and evaluated within each tissue type.
The following figure illustrates the exact methodology of querying GPt-4 for annotation generation.
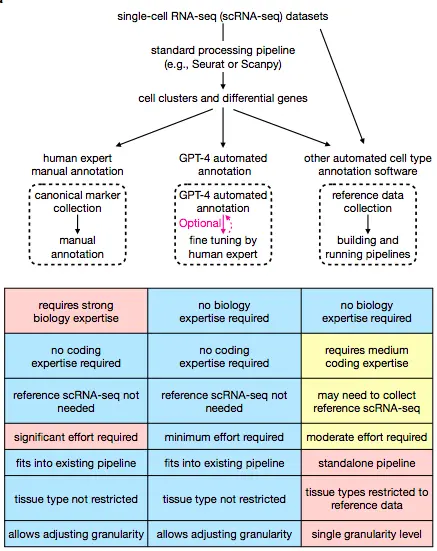
Image source: https://www.biorxiv.org/content/10.1101/2023.04.16.537094v1.full.pdf
How Well did GPT-4 do, Really?
- In all the annotation tasks performed, GPT-4’s annotations match fully or partially with manual annotations for at least 75% of cell types.
- The authors grouped cell types into major cell categories as per manual annotations, and the concordance between GPT-4 and manual annotation was found to be the greatest for homogeneous cell categories and lowest among heterogeneous categories such as in stromal cells.
- When tested for identifying cell clusters comprising a mixture of cell types on simulated datasets with canonical markers from different cell types, GPT-4 was able to discriminate between single and mixed cell types with an average accuracy of 94%.
- GPT-4 exhibits robust performance in data analysis scenarios and has been shown to be 100% accurate in differentiating between known and unknown cell types, on average.
Conclusion
Manual cell-type annotation has been a cumbersome process in the scRNA-seq analysis pipelines. The advent of automated annotation pipelines provided some relief, however, these require additional pipeline developmental efforts and are tuned for specific reference datasets. The authors in the current article, available in bioRxiv, have demonstrated the usability of GPT-4, the most advanced large language model for automating the cell-type annotation step. The results are truly encouraging, and the authors envision that the manual annotation efforts will soon be replaced by the chatbot-aided annotation methodology, a truly remarkable feat for the deep learning paradigm in the realm of the life sciences.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.










