In a recently published study, Stanford Medicine researchers highlighted age-related problems in humans and showed how aging varies from person to person and between organs within an individual. Researchers used human blood plasma protein levels from particular organs to examine aging in 11 key organs in real people. The approach suggested by the authors offers a straightforward and comprehensible way to research the aging of organs. Understanding the molecular changes in human organs is crucial for addressing the global disease burden and revolutionizing patient care, preventative medicine, and drug development. Current methods present are either expensive or need to provide the required molecular insights. To combat this problem, the researchers hypothesized that complete quantification of organ-specific proteins in plasma could enable minimally invasive assessment and tracking of human aging for any organ.
Key Findings
- 1.7% of people show aging in multiple organs, and nearly 20% exhibit significantly accelerated aging in one organ.
- There is a 20–50% increased risk of death associated with accelerated organ aging, and some disorders are associated with accelerated organ aging.
- Individuals with accelerated heart aging have a 250% higher risk of heart failure, and accelerated brain and vascular aging predict the course of Alzheimer’s disease (AD) just as well as plasma pTau-181, the most effective blood-based biomarker for AD at present.
We can estimate the biological age of an organ in an apparently healthy person
– Tony Wyss-Coray
Decoding the Plasma Proteome: A Gateway to Aging Insights
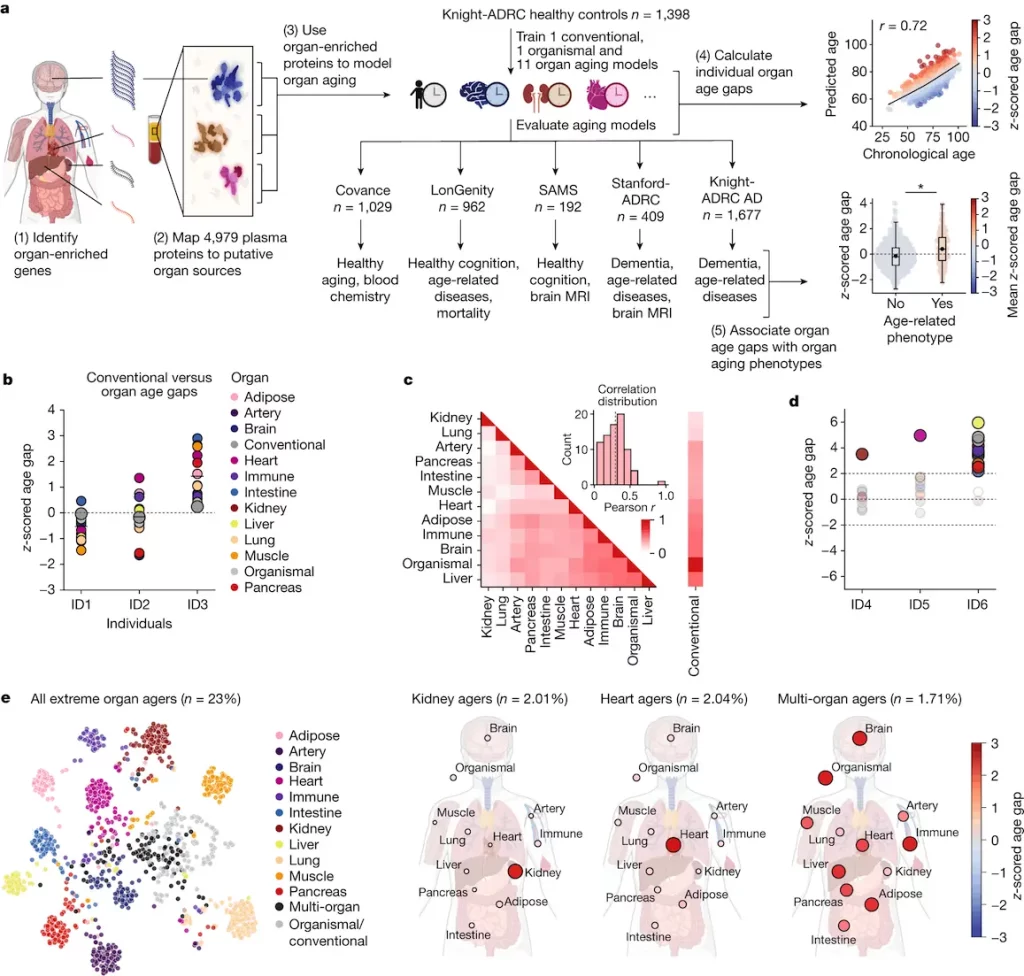
The study mapped the organ-specific plasma proteome using RNA-seq data from the Genotype-Tissue Expression project. The SomaScan assay measured 4,979 proteins across 5,676 subjects, with 18% of them being ‘organ enriched'(The Human Protein Atlas provided a definition for “organ enriched” genes, which were defined as those expressed at least four times greater in one organ than in any other organ). The highest number was found in the brain, with the highest number from the brain for theSomaScan assay. Quality control removed high-variation or low-correlation proteins, leaving 4,778 proteins (17.9%) for downstream analysis.
Adipose tissue, arteries, brain, heart, immunological tissue, gut, kidney, liver, lung, muscle, and pancreas were among the 11 major organs for which the study trained a bagged ensemble of least absolute shrinkage and selection operator (LASSO) aging models. The Knight Alzheimer’s Disease Research Centre group comprised 1,398 healthy individuals who underwent testing of the models. All five cohorts’ ages were significantly estimated by the organismal model and all 11 organ aging models following multiple test corrections. The method produced highly enriched proteins specific to particular organs and had specified activities.
At the population level, individuals with the same conventional age gap had diverse organ aging profiles, with a low-to-moderate correlation between age gaps of different organs (mean pairwise Pearson r = 0.29). However, the majority of variance in one organ age gap is not explained by others, except for organismal and conventional age gaps, which were highly correlated. Some individuals had extreme aging in one or more organs, suggesting that organ age gaps may capture unique aging information.
Unveiling Organ Aging Signatures
The study examined the connection between biological aging and organ age in two cohorts. The most significant correlations were discovered between kidney aging and metabolic disorders, cardiac conditions (heart attack and atrial fibrillation), muscle aging and impaired gait, brain aging and cerebrovascular illness, and organismal aging and Alzheimer’s disease. While atrial fibrillation and cardiac attacks were associated with heart aging attributes, the kidney age difference was linked to characteristics of metabolic diseases. The study found that kidney enzyme renin (REN) and kidney-associated antigen 1 (KAAG1), as well as uromodulin (UMOD), are key kidney aging proteins. Chronic kidney disease has been genetically related to UMOD, and autosomal dominant tubulointerstitial kidney disease is primarily caused by uncommon mutations.
The present study highlights the possibility of a connection between “normal” heart aging and subclinical heart illness, which warrants additional research using more precise heart imaging and electrophysiology. After multiple test corrections, the study demonstrated that, after 15 years of follow-up in the LonGenity cohort, the age gaps from 10 out of 11 organs, the conventional model, and the organism model were substantially linked with future risk of all-cause mortality.
The Covance project conducted in this study investigated the connection between clinical markers, such as phenotypic age (PhenoAge), and organ age disparities. The findings indicated a strong link between various organ age differences, with other factors accounting for a very modest percentage of the variance.
Navigating the Rhythms of Brain Aging in Cognitive Decline
Researchers from the Stanford Alzheimer’s Disease Research Centre (Stanford-ADRC) and the Knight-ADRC cohort have examined the brain age gap, a major risk factor for neurodegenerative illnesses. The objective of the feature importance for biological aging (FIBA) algorithm is to comprehend the role played by underlying proteins in the brain aging model’s capacity to forecast brain aging phenotypes. This work used the information from the above studies to build the CognitionBrain model.
In the Knight-ADRC cohort, the CognitionBrain aging model—which was trained using brain-specific proteins called CDRGLOB FIBA+—had a greater correlation with AD than the conventional age gap and the first-generation brain age gap. In all ADRC cohorts, the model was also substantially linked to the probability of dementia development in the future. It has also been discovered that the CognitionBrain age difference predicts brain volume in a number of AD-sensitive areas. To more accurately stratify AD patients for potential clinical outcomes, the model may be utilized in conjunction with additional biomarkers of AD and indicators of cognitive loss. Molecular information about brain aging that is not captured by other methods is provided by the CognitionBrain age gap.
Additionally, through the proteins tenascin R (TNR), neurocan (NCAN), and heparan sulfate-glucosamine 3-sulfotransferase 4 (HS3ST4), the model implicated changes in the glycosylated extracellular matrix, highlighting the extracellular matrix’s function in brain aging.
In the Knight-ADRC and Stanford-ADRC cohorts, the study evaluated the highest weighted CognitionBrain proteins for changes with age and AD, as well as changes with AD in brain tissue at the protein, levels of bulk and single-cell RNA from databases that are accessible to the public. With age and AD, a regular trend of blood volume rising and AD brain tissue shrinking is observed. This implies that a loss or change in protein processing and subsequent shedding of these essential components in the brain may be reflected in the elevation of synapse and neurite development associated with protein levels in the blood.
Future Directions
Future studies may evaluate the correlation between proteomic organ aging and additional molecular indicators of aging and illness, like methylation aging clocks and disease-specific prediction models. The technique is platform-neutral, but the present models rely on about 5,000 proteins detected using the SomaScan test. The increasing quantity of single-cell resolution human gene expression maps will aid in the improvement of organ and cell-type specific aging models and enable a thorough comprehension of organismal physiology based on the plasma proteome.
Conclusion
The study presents a model for employing plasma proteins to estimate biological aging and organ health. The developed models are predictive of aging heterogeneity amongst tissues, illness risk and progression, and organ-specific functional decline. This method can be used to comprehend how health interventions affect individual organs. The researchers calculated the organ ages of any plasma proteomics sample using the SomaScan assay and an intuitive Python tool called organage.
Article Source: Reference Paper | Reference Article
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.