Preimplantation genetic testing (PGT) of embryos fertilized in-vitro has been proposed as a strategy to diminish transmission of common diseases; in any case, a more comprehensive embryo genetic assessment, joining the impacts of common variants and rare variants, stays inaccessible.
In this study, the scientists used molecular and statistical techniques to infer inherited genome sequence in 110 embryos and model susceptibility across 12 common conditions.
The scientists noticed a genotype accuracy of 99.0-99.4% at sites relevant to polygenic risk scoring in cases from day-5 embryo biopsies and 97.2-99.1% in cases from day-3 embryo biopsies.
Joining rare variants with a polygenic risk score (PRS) amplifies predicted differences across sibling embryos. For instance, in a couple with a pathogenic BRCA1 variant, the scientists predicted a 15-fold distinction in odds ratio (OR) across siblings while combining versus a 4.5-fold or 3-fold contrast with BRCA1 or PRS alone. Their findings might illuminate the discussion of the utility and implementation of genome-based PGT in clinical practice.
The Preimplantation Genetic Testing (PGT)
The preimplantation genetic testing or PGT empowers the profiling of embryos for family-specific genetic disorders before implantation. Even though PGT is utilized in forestalling rare Mendelian disorders, a few groups have investigated expanding testing to include common conditions, like heart disease and cancers.
These methodologies depend on the utilization of a PRS7 that consolidates the effects of tens or hundreds, or thousands of genetic variants into a single predictor. In any case, due to the limited quantity and quality of DNA in single-cell or few-cell embryo biopsies, endeavors to comprehensively profile the genomes of embryos are expensive and time-intensive, experience the ill effects of inaccuracies connected with allele dropout, require expanded relatives or on the other hand depend on imputation, which hampers the detection of rare deleterious variants in genes like BRCA1.
To conquer these limits, the scientists extended a strategy for whole-genome reconstruction (WGR) that utilizes parental genome sequences and embryo genotyping to predict the inherited genome sequence of an embryo. They applied this approach to deal with 110 embryos across ten couples and compute polygenic predictors across 12 medical conditions, including cancers, cardio-metabolic and autoimmune diseases. The reconstructed genome and polygenic predictions for the embryos were compared to those produced from a tissue sample of the corresponding born children.
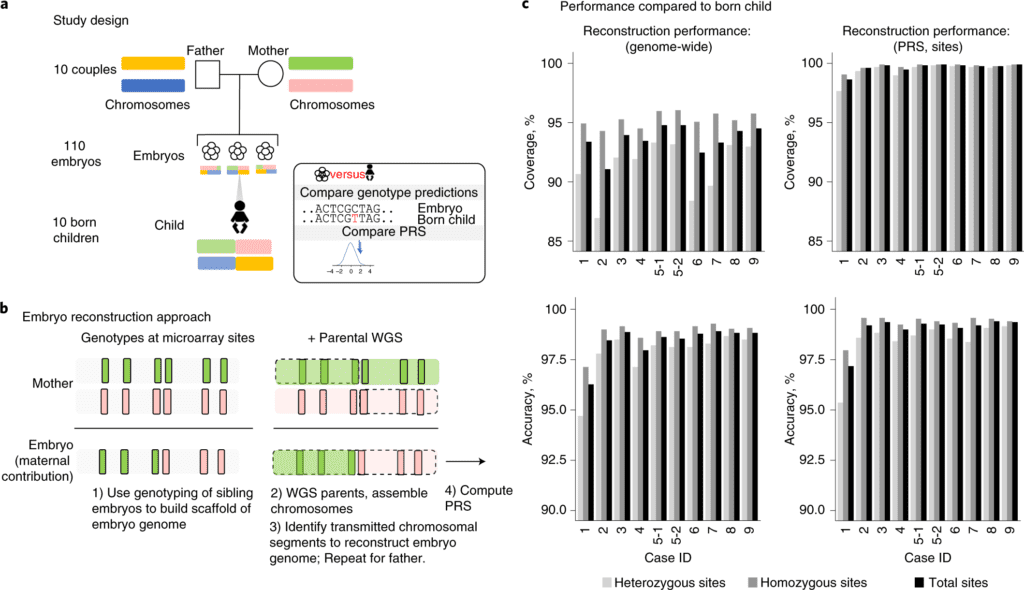
Image Source: Whole-genome risk prediction of common diseases in human preimplantation embryos
Scopes of Improvement
Several areas of improvement remain in the preclinical research. Some of them that are extensively discussed in the paper are as below:
- Limitation to Inherited Genetic Variation
Even though the scientists affirmed inheritance at a few billion sites in a born kid, they barred variants that might be ‘new’ or de novo in the gametes or somatic mutations that emerge after conception. Although rare, de novo variants represent a significant part of specific early-stage neurodevelopmental disorders, including autism and intellectual disability.
Their predictions were more precise in day-5 embryos thought about with day-3 embryos, logical because of increased cellular material present in a trophectoderm biopsy. As WGS costs decline, they anticipated their methodology could be supplemented with ultradeep sequencing of embryo biopsies to identify these variants.
- Limited Effectiveness of PRSs in Non-European Populations
Human genetics research historically involved elaborate members of European ancestries, and thus, the predictive power in non-European cohorts is restricted. In the long haul, procedures including recruitment of ancestrally diverse cohorts and fine-mapping to the causative variants should further develop performance across ancestries. Temporarily, utilizing a subset of models with evidence of cross-ancestry performance might be fundamental.
- Using PRS for Clinical Utility for Reduction of Disease Risk in Preimplantation Embryos
Discoveries from research cohorts may not sum up to sibling embryos in IVF and diminish their prediction accuracy. These biobanks are predominantly made out of unrelated individuals; a few gatherings reported a decline in the predictive ability of models trained to utilize unrelated individuals inside families, especially for cognitive and psychiatric traits.
The analysis of a cohort of siblings versus unrelated individuals in the UKB, utilized to generate the respective OR among sibling embryos, brought about comparative breast cancer PRS effect sizes for siblings versus unrelated individuals. More family-based prospective studies of genetics and disease are expected to reduce these concerns.
Besides, autonomous of the intrafamily effect, children born at present might face a different environment and risk profile than grown-up members in most research biobanks. Independently, people with a family background of disease might display a higher absolute risk reduction of disease because of a higher reduced chance of disease.
- Incorporating Polygenic Information into Prenatal Decision-Making may raise Ethical and Practical Questions that deserve much Consideration in the Field.
Higher is the risk of unequal access to this innovation for families because of either the expense of IVF or constraints in the cross-ancestry performance of polygenic models. The studies carried out by the scientists are free to participants and include several models validated across multiple ancestries. Nonetheless, widespread disparities should keep on being tended to.
Regardless of limits, emerging evidence recommends public interest in combining PRS in embryo screening; a recent survey found 68% of 1,457 US members accepted embryo screening utilizing PRS was reasonable. Further review, through research conventions like our own, can assist with producing truly necessary evidence on utility.
Intended Discussion
The scientists utilized a combination of molecular and statistical approaches to link parental variants into ‘haplotypes’ corresponding to individual euploid chromosomes, determine sites of meiotic recombination for every embryo, and assemble the relevant haplotype segments between every one of the recombination sites to approximate the whole inherited embryonic genome.
They assessed the precision of WGR by comparing their actual genotype to the actual genotypes present in the born children. The scientists anticipated an average of 5.8 million sites in embryos, going from 5.4 to 6.4 million variants. Genome-wide prediction accuracy went from 96.3-98.4% in day-3 embryo biopsies to 98.0-98.9% in day-5 embryo biopsies compared to the born kid.
They inferred that processing and testing of single cells from day-3 blastomere biopsies yields decreased performance compared to processing and testing of multiple cells from day-5 trophectoderm biopsies.
The Endpoint
In this preclinical research, the scientists exhibited that WGR for embryos is achievable and empowers accurate calculation of PRS when compared with results from genomic samples from 10 born children. They noticed heterogeneity in predicted disease risk across families and siblings. PRS on numerous common variants with small effect sizes, combined with rare variants, might better capture the heterogeneity in predicted disease risk.
As our understanding and the predictive power of PRSs gets to the next level also, sequencing technology turns out to be more cost-effective. This approach might be utilized to dependably infer inherited genome sequence. Also, the model predicted genetic risk in embryos of couples with a personal and/or family history of common disease undergoing IVF.
Story Source: Kumar, A., Im, K., Banjevic, M. et al. Whole-genome risk prediction of common diseases in human preimplantation embryos. Nat Med 28, 513–516 (2022). https://doi.org/10.1038/s41591-022-01735-0
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.