Researchers from the Department of Biological Chemistry at Hebrew University, Jerusalem, and the Department of Pharmaceutical Chemistry at the University of California, San Francisco, have developed a landmark tool to forecast how mutations influence protein interactions. This study by Dr. Julia M Shifman, Dr. William DeGrado, and the team describes ProBASS, a language model that can predict changes in binding affinity due to mutations.
Understanding the Importance of Protein Interactions
Proteins are the workhorses within our cells as they perform numerous tasks. They interact extensively to perform cellular functions. A single alteration in a protein’s sequence, known as mutation, can have drastic effects on these interactions. Understanding how mutations affect protein binding is essential for decoding disease mechanisms and designing new drugs.
Traditionally, predicting the impact of mutations on protein binding has been challenging. On the one hand, experimental methods take a lot of time and money, while on the other hand, earlier computational methods were not precise enough. However, with recent improvements in artificial intelligence, particularly in language models, it is now possible to obtain new possibilities.
How ProBASS Works?
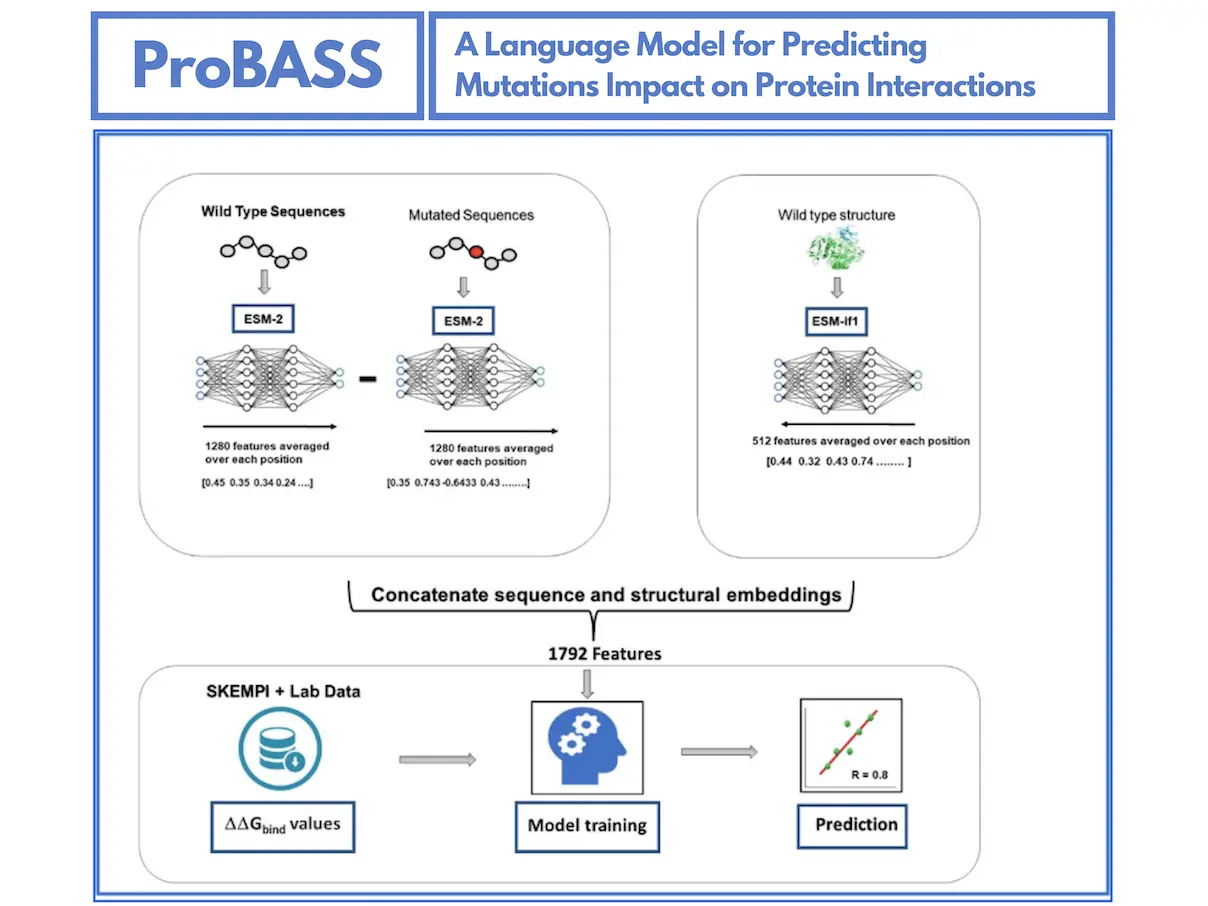
ProBASS is a powerful tool that utilizes Protein Language Models (PLMs) to forecast how mutations will affect protein-protein interactions. The PLMs are sophisticated programs trained on large amounts of data on protein sequence and structure. There are two of the most advanced PLMs used in ProBASS:
- ESM-2: Focuses on the proteins’ sequences to give information about the order of amino acids.
- ESM-IF1: Concentrates on the proteins’ structures to understand the three-dimensional arrangements of amino acids.
By blending these two models, ProBASS can make an all-embracing depiction of a protein-protein complex. Whenever a mutation occurs, it recalibrates these representations to predict how such mutations alter binding affinity.
A Leap Forward in Prediction Accuracy
Researchers trained Probass using an enormous database containing different experimental data on binding affinities across several protein-protein interactions. For single and double mutations alike, it was highly accurate in forecasting their effects on binding affinities.
Complex protein-protein interactions can be handled by ProBASS, which is one of its key strengths. Several mutations were difficult to predict with the existing methods; nonetheless, ProBASS can estimate double mutations.
A Broader Approach Than Single Mutations
What is particularly remarkable about ProBASS is its ability to deal with not only single but also double mutation cases. The significance of this development is that there are many diseases resulting from more than one genetic alteration, and taking into account the combined actions of two mutations will help better understand potential consequences.
The model has learned complex patterns and relationships between protein sequences, structures, and binding energies through its training on an extensive dataset that includes experimental binding affinities. This high level of learning has resulted in enormous predictability, surpassing other methods.
The Potential Impact
Protein interactions are now better understood and manipulated thanks to the progress of ProBASS. This means that many fields/areas can be positively affected by this tool, such as:
Drug discovery: To efficiently determine proteins that interact with other proteins causing certain diseases, researchers should anticipate how various mutations may affect these PPIs.
Protein engineering: Using specific mutations’ impact on protein properties, ProBASS can help design proteins with desirable features.
Basic research: It is through comprehending these fundamentals of biology that every aspect related to protein-protein interactions could be addressed. In this regard, ProBASS will provide information about mutated impacts.
Looking ahead: Challenges and Opportunities
Nonetheless, ProBASS is greatly progressive and, thus, still has opportunities to improve. By including a larger number of experimental data types and expanding the training dataset, the accuracy of the model and its generalizability may be significantly improved.
Furthermore, effective methods of interpreting model predictions are required. Once we know what makes the model think, it will provide new insights into protein-protein interaction mechanisms that could be used for targeted interventions.
ProBASS is an example of some computational biology tools that are leading in this fast-growing field. Other more advanced models using much more sophisticated algorithms are likely to emerge as technology advances further in solving biological systems complications.
Join the Conversation!
You are welcome to post your contribution on how ProBASS can affect different spheres. Feel free to share any thoughts you have or ask questions related to it to let us examine multiple perspectives!
We look forward to hearing from you!
Article Source: Reference Paper | Software could be downloaded from GitHub.
Important Note: BioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.
















[…] ProBASS: A Game-changer in Predicting Mutation Impact on Protein Interactions […]