Scientists from Case Western Reserve University, Cleveland, USA, developed a self-supervised image representation learning framework, “ImageMol,” for predicting molecular properties and drug targets with high accuracy, making it a valuable tool for computational drug discovery.
Imagine we belong to the incredibly lazy bunch that uses a pre-made pizza crust that would probably make every Italian and hardcore pizza lover out there appalled. Though, despite its controversial existence, nobody can question its functionality. It’s fast and efficient based on your choice of topping, it’s easily customizable, and at the end of the day, you still got a pretty decent pizza. Likewise, or even better is the existence of ImageMol. It’s fast, efficient, and easily customizable to your choice of drug discovery process, and at the end of the day, you get a highly accurate prediction model.
Why the need for ImageMol?
C4H10 can go from being a fuel for cigarette lighters with a boiling temperature of 1°C (butane) to a refrigerant with a boiling temperature of -11°C (isobutane) with a slight change in its structure from linear to branched. This was, but an exercise for stressing the extreme influence the structure of a molecule exerts on its properties and hence its functions. Suppose one could gauge the properties of a drug from its structure alone. In that case, it can prove highly beneficial in studying the reactions and interactions of the said drug in different scenarios or, in other words, its pharmacokinetic and pharmacodynamic properties within a biological system. It might be a tedious, time- and resource-consuming task for a human but a cakewalk for an adequately trained computer model. And luckily enough, we have a good deal of those in our midst, and more and more are entering this highly competitive platform.
ImageMol, a pre-trained deep learning framework for computational drug discovery, accepts an input dataset of molecular structures in the form of images labeled with the target variable for prediction. The model then identifies global and local structural characteristics important for the given prediction task. And like the range of pizza toppings, we could go crazy on the possible drug discovery datasets that could be applied to this model. The authors themselves were able to train the model to perform a variety of classification and regression tasks, from predicting the drug’s molecular properties (metabolism, toxicity, solubility, blood-brain barrier penetration) to classifying potential drug candidates against human immunodeficiency virus (HIV) and beta-secretase for Alzheimer’s disease to assessing the binding activities at kinases and G protein-coupled receptors all with high accuracy outputs. These diverse studies were performed using benchmark datasets with experimentally obtained values to train the ImageMol framework initially. Not bad for a pre-made crust, is it?
How does ImageMol work?
The context behind pre-trained models is to utilize the previously acquired knowledge in solving newer problems. It involves injecting the current datasets into the architecture and weights of the previously designed neural models pre-trained on large datasets with similar data, thus skipping a lot of the initial steps in developing a model completely from scratch.
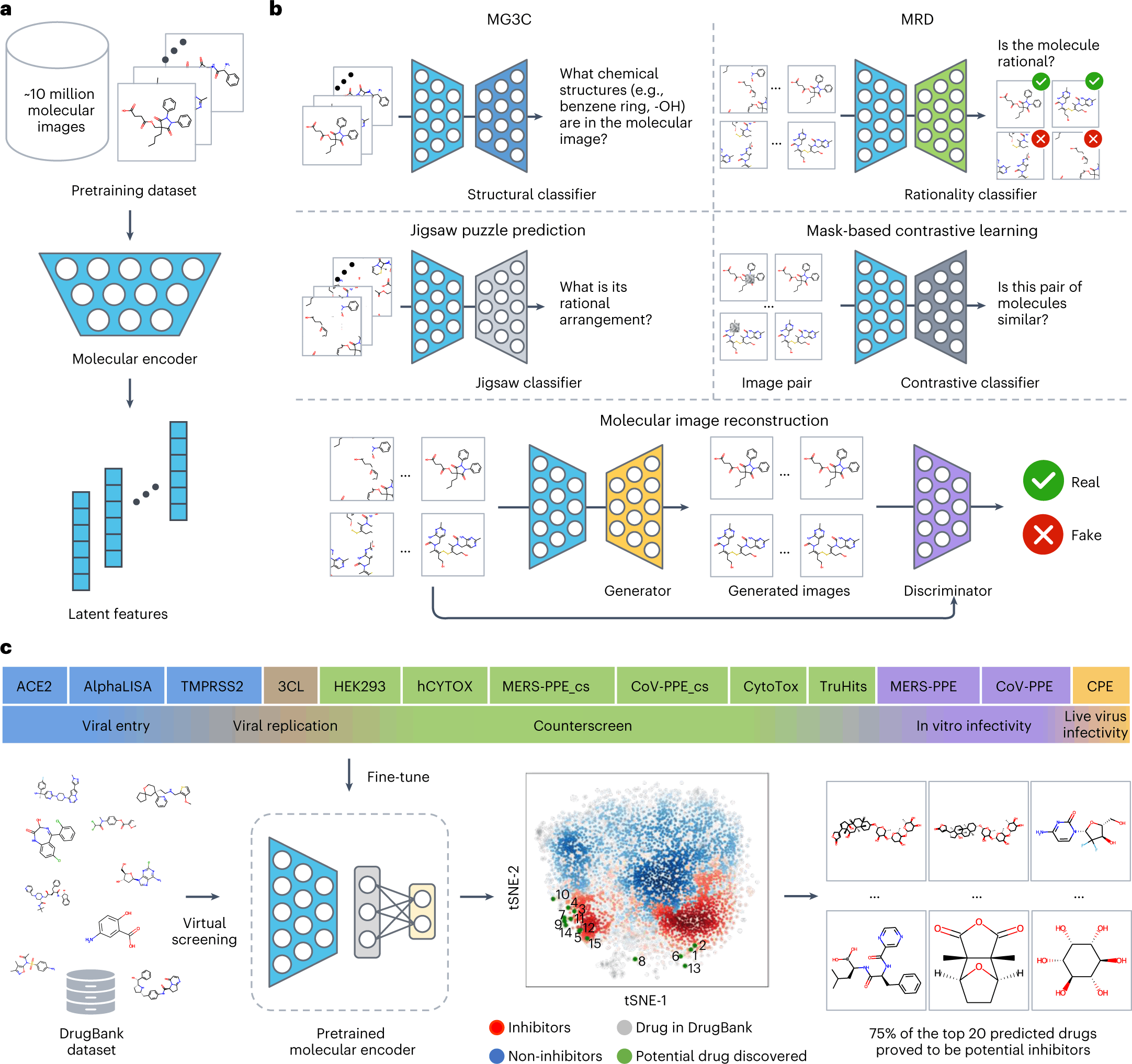
ImageMol itself makes use of ResNet18, an image recognition convolutional neural network that is pre-trained to classify images into more than 1000 objects, as the molecular encoder to derive the latent features. But in order to attune the molecular encoder to the task of deriving structural features from molecular images, the model was first trained on the following three principles: Consistency, Relevance, and Rationality. Five pretext tasks(given in bold) were then defined to assess how well the model implements the said principles.
- Consistency: To study if different images of the same chemical structures are extracting consistent structural features.
Multi-granularity chemical clusters classification (MG3C): Clusters of different granularity(structural details) were obtained through K-means clustering of the MACCS fingerprint for each molecule, with each of the clusters being assigned as a pseudolabel. The pseudolabels for each molecule were compared with the predicted labels from the model for all the molecular images of the same structure. The model consistency was improved by optimizing the loss between the predicted labels and the pseudolabels.
- Relevance: To study whether different augmentations of the same chemical structure are related or not. Two strategies were adopted for implementing this:
Molecular image reconstruction (MIR): The model was first fed with the correct image and then with a shuffled and rearranged version of the same. The model was assessed based on its ability to reconstruct the original image by comparing features extracted from both and classifying the reconstructed image as real or fake.
Mask-based contrastive learning (MCL): A 16×16 square area of the molecular image is masked. The model is trained to minimize the distance between the latent features extracted from masked and unmasked images.
- Rationality: To study the rationality of the molecular structures in the images.
The images were decomposed into nine patches, and the patches were rearranged.
Molecular rationality discrimination (MRD): Predicted whether the input image was rational or irrational.
Jigsaw puzzle prediction (JPP): Assigns labels to each patch and randomly shuffles the patches to study if the model could predict the correct order.
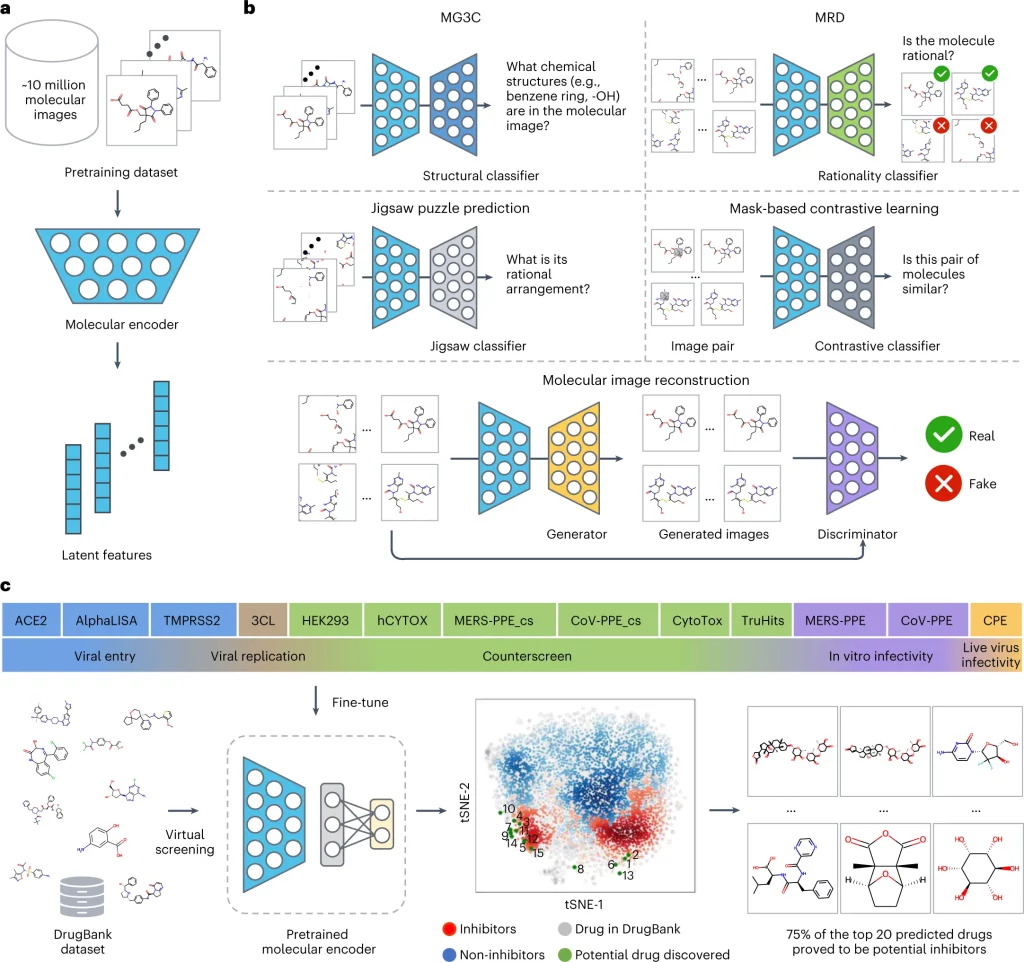
Image Source: https://doi.org/10.1038/s42256-022-00557-6
The pre-training process of ImageMol used 9,999,918 structures of drug-like bioactive molecules from Pubchem that are first augmented by three methods to improve the model generalizability: RandomHorizontalFlip, RandomGrayscale, and RandomRotation. The forward propagation strategy of Imagemol involves passing these images through the Resnet18 molecular encoder that derives their latent structural features. These features act as inputs for the above five pretext tasks, and the loss is calculated as:
LALL = LMG3C + LJPP + LMIR + LMRD + LMCL
The total loss is then backpropagated to update the ResNet18 through a stochastic Gradient descent algorithm.
A pre-trained model is usually finetuned to the specific task at hand before applying it to unlabelled data. In this particular study, though, the focus is on the model development, and they have added naught but an additional output layer after ResNet18, whose dimensions are on par with the downstream task. Even in the absence of a more intricate finetuning, the model performed robustly, often times outperforming other baseline models for the same task. The authors have taken the study a notch higher and have successfully applied the model on high-throughput experimental datasets from NCATS (National Center for Advancing Translational Sciences) to identify 3-C like protease inhibitors against SARS-Cov2.
Where to next?
Though the current version is remarkable by itself, the authors have acknowledged shortcomings and potential directions to improve the model further. They have hinted at ImageMol version 2.0 addressing some of these concerns, such as the integration of 3D information and atomic properties crucial for predicting molecular properties and drug interactions.
Conclusion
Overall, ImageMol is a self-supervised image processing-based approach that offers a comprehensive set of tools for computational drug discovery. The method is appropriate for identifying new drug targets and developing new therapeutics for various human diseases, as well as COVID-19.
Article Source: Reference Article | ImageMol
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Catherene Tomy is a consulting Content Writing Intern at the Centre of Bioinformatics Research and Technology (CBIRT). She has a master’s degree in Molecular Medicine from Amrita University with research experience in the fields of bioinformatics, cell biology, and molecular biology. She loves to pull apart complex concepts and weave a story around them.
.