Recent achievements in the field of artificial intelligence (AI) have helped to develop effective medical imaging frameworks that can achieve professional clinical expertise. However, they often generated inadequate outputs while assessing clinical data, which was different from the training data. Google researchers and collaborators at the Georgia Institute of Technology and Northwestern University have proposed a new medical imaging framework, REMEDIS, that can analyze complex medical imaging data using universal supervised transfer-based algorithms with self-supervised learning.
AI-based techniques are already in use for medical imaging. They are dependable, economical, and take less time to complete extremely difficult tasks. They create individual systems for various clinical settings based on site-specific data. However, this approach is inefficient because of the high volume and diversity of medical data. In order to create an effective medical AI platform, clinical data needs to be generalized.
Generalization remains a key translational challenge for medical imaging applications. AI systems are evaluated based on in-distribution (ID) or out-of-distribution (OOD) settings. In an in-distribution (ID) setting, the AI system uses data as the trained model, whereas an out-of-distribution (OOD) uses a relatively new clinical setting from the dataset used for training machine learning models. AI models for medical imaging were observed to perform better in ID settings than in OOD settings.
Data-Efficient Generalization
A system was proposed earlier to design new AI models for new data to overcome the problem related to the generalization of variable clinical datasets. The approach was rejected because it was impractical and expensive, as it takes time to collect and analyze clinical data.
A group of researchers, software engineers, and clinicians from Google have introduced a new approach to tackle this problem. Data-efficient generalization was achieved by:
1) Observing the OOD setting’s performance with restricted access to training data. The process was termed “zero-shot generalization” and
2) Reducing the dependency on training data from OOD settings to achieve clinical-level expertise.
The Development of Self-Supervised Algorithms for Creating AI Systems.
The two biggest challenges faced by researchers while designing AI systems for medical data imaging are acquiring labeled data and reducing dependence on training data. Self-supervised learning approaches used in medical AI settings reduce the dependence on labeled data while devising AI-based models. They use pre-text functions to train machine learning algorithms without using labeled data.
The ability of self-supervised learning methodologies to lessen the reliance on labeled data while developing AI-based models has increased their use in medical AI settings. Contrastive learning is a self-supervised learning method that involves training the model to differentiate between similar and different pairs of images or data points. Generative modeling is another approach that trains the model to generate new examples from the existing unlabeled data. This approach can be mainly helpful in situations where labeled data is limited or expensive to acquire.
Data-Efficient Generalization Using Self-Supervision Algorithms
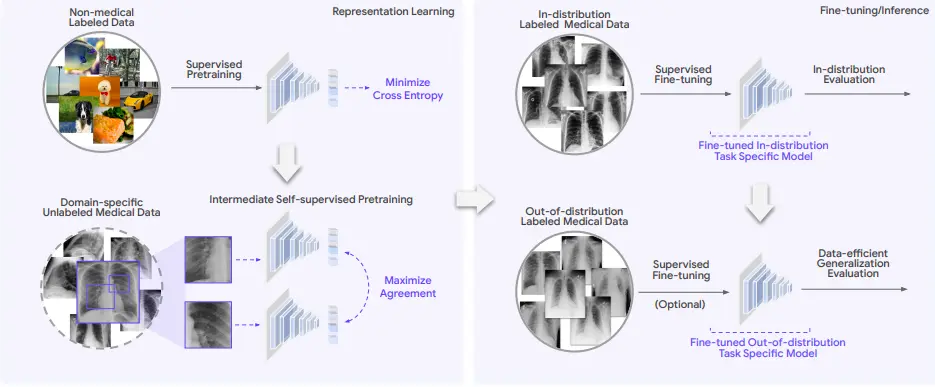
REMEDIS stands for “Robust and Efficient Medical Imaging with Self-Supervision.” It uses a self-learning-based machine-learning approach to perform data-efficient generalization. It efficiently performed AI-based clinical imaging with minimal customization. Both generalized medical images and non-medical images are used to train the algorithm. The REMEDIS platform integrates both large-scale as well as task-specific supervised learning algorithms using less labeled data to carry out medical imaging tasks.
The REMEDIS method can be summarized as follows:
- Pretraining a medical AI model by a supervised learning algorithm based on labeled non-medical images using the Big Transfer (BiT) method.
- Using a self-supervised learning algorithm to achieve data-efficient generalization.
The REMEDIS approach uses extensive supervised pretraining data and intermediate self-supervised learning algorithms to be used across different medical imaging modalities without needing domain-specific customization. The REMEDIS approach depends on learning based on unlabeled data using contrastive self-supervision. This method is suited for medical imaging AI due to the lack of expert-labeled data and the relative abundance of unlabeled medical images.
The REMEDIS approach aims to develop predictors for each medical task that are specific to a particular domain and have a low prediction error on both in-distribution and out-of-distribution data. Predictors use these pre-trained representations and are further fine-tuned using the labeled data. Pretraining on unlabeled datasets may improve accuracy under distribution shifts.
It has been discovered that using supervised pre-trained models enhanced downstream task performance. Utilizing modern training methods like Big Transfer (BiT) and alterations to the current algorithm was helpful in accelerating training and enhancing downstream task performance.
Clinical Evaluation and Validation
To evaluate REMEDIS, six tasks across five medical imaging modalities were performed, as mentioned in the article posted on arXiv. The five selected medical imaging modalities were: chest radiography, mammography, clinical dermatology photography, digital pathology, and fundus imaging. REMEDIS and baseline AI models were tested against previously unseen datasets from a different clinical setting that was used to train a different AI system. Each specific modality and task included an unlabeled pretraining dataset, an ID dataset, and one or more OOD datasets. Unseen datasets from a different clinical environment were used to train REMEDIS and baseline AI models.
An example: A comparative study was carried out to examine REMEDIS’s usefulness for classifying dermatological conditions. A pre-existing AI model trained to identify various skin conditions based on images taken with a digital camera was chosen. The result showed REMEDIS efficiently reduced the usage of labeled data along with efficient learning capabilities.
Limitations and Future Work
REMEDIS was designed using a wide range of tasks and medical imaging domains. Learning strategies like self-training, weak supervision, and active training have been left out while designing REMEDIS. Being a relatively new field of AI, self-supervised learning algorithms can be challenging to use effectively for a given task, especially for researchers and developers with low computing power.
Conclusion
REMEDIS significantly reduced the amount of labeled data needed for new deployment sites. By addressing a number of important issues, such as the speed of obtaining local data, which may take years to collect, REMEDIS was able to prove its efficiency. Moreover, monitoring the ethics, privacy, and fairness of using self-supervised learning on large amounts of data to generate medical AI is an important area for future research.
Article Source: Reference Paper | Reference Article
Learn More:
Sipra Das is a consulting scientific content writing intern at CBIRT who specializes in the field of Proteomics-related content writing. With a passion for scientific writing, she has accumulated 8 years of experience in this domain. She holds a Master's degree in Bioinformatics and has completed an internship at the esteemed NIMHANS in Bangalore. She brings a unique combination of scientific expertise and writing prowess to her work, delivering high-quality content that is both informative and engaging.