Scientists from Xi’an Children’s Hospital have shown that metagenomic next-generation sequencing (mNGS) of cell-free DNA (cfDNA) and RNA may help detect trace infections from various bodily fluids and blood samples extracted from newborns. Researchers also performed a local polynomial regression fitting analysis, which revealed that the best time for detecting mNGS was 1 to 3 days following the initiation of antimicrobial medication. The study’s findings emphasize the value of mNGS in detecting causative DNA and RNA infections in infected neonates.
Newborns are frequently susceptible to infections, such as sepsis, meningitis, and pneumonia, which account for millions of infant deaths yearly. Apart from the mortality rates, adverse prognoses may arise in neurodevelopmental problems and long-term disability if timely treatment is not given. Pathogenic microorganisms causing such infections include a diverse group of bacteria, viruses, and fungi. Therefore, timely and valid pathogen identification and appropriate therapeutics are required to enhance prognoses.
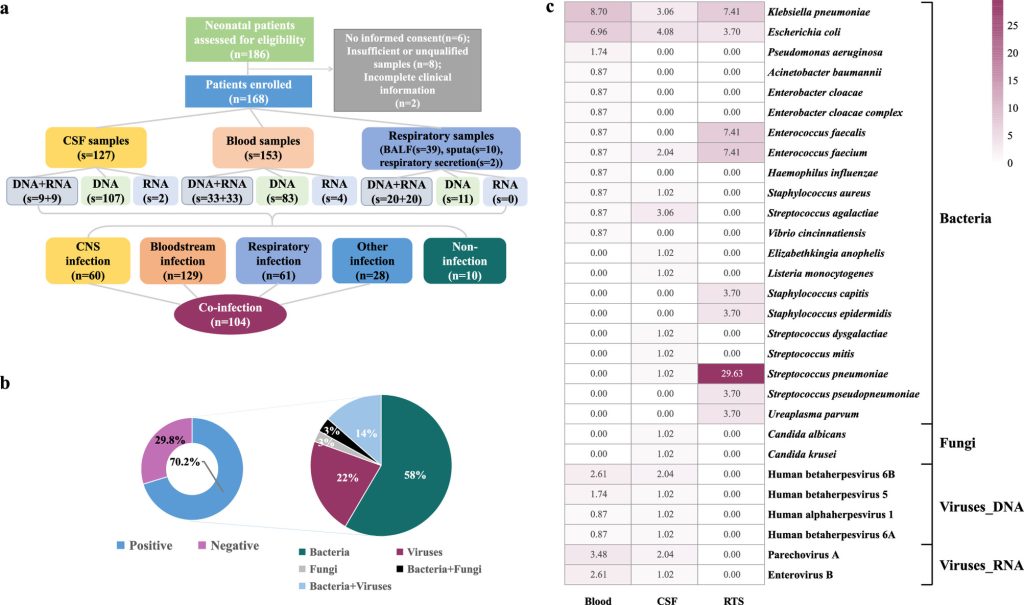
Image Source: https://doi.org/10.1128/spectrum.01195-22
Blood, cerebrospinal fluid, etc., serve as excellent samples for newborn infections. However, more than 60% of illnesses are undetectable in blood or cerebrospinal fluid (CSF) cultures. Although using PCR or antibody techniques can help detect a small number of pathogens in an experiment, metagenomics next-generation sequencing (mNGS) in clinical infections of the central nervous system (CNS) promises to be an unbiased technology to detect pathogens in a variety of diseases. Metagenomic next-generation sequencing runs all nucleic acids in a sample, which may contain a mix of microorganism populations.
Thus, the present study by Chen et al. collected neonatal fluid samples with suspected illnesses to compare the accuracy of mNGS using cfDNA and RNA to conventional methods followed for clinical diagnosis. A regression model was also devised to assess the impact of therapy on pathogen detection using mNGS and traditional methods and to predict the ideal time for mNGS testing.
Major Findings
This study seeks to be the first to evaluate the performance of mNGS in diagnosing newborn infections using cfDNA and RNA from body fluid and blood samples. In DNA and RNA assays, mNGS outperformed conventional procedures, with a high total coincidence rate (TCR) of 68.3%. The significant pathogens in blood samples, CSF samples, and other samples were K. pneumoniae, human beta herpesvirus 5, E. coli, and S. pneumoniae. Given the inevitability and necessity of empirical therapy, the ideal time for mNGS testing was found to be one to three days following the initiation of antimicrobial medication, and mNGS plays an important role in providing a reference for clinical care. The high TCR results confirmed that mNGS was superior to conventional methods for pathogen detection using blood and body fluid samples.
Most newborns in this study (104 out of 168 samples) had various coinfections. Some microbes, such as S. pneumoniae, can spread throughout the body, from the upper respiratory tract to other organs, including the meninges, via the bloodstream. In the study, E. coli was found in blood and CSF samples from patients with CNS-bloodstream coinfections, whereas S. agalactiae was found only in CSF samples. The continuous use of antimicrobial drugs may result in a much lower load of S. agalactiae in the blood than that of E. coli. The findings highlight the importance of metagenomic next-generation sequencing testing in blood and CSF for diagnosing neonatal CNS-bloodstream coinfections. However, some microbes, such as cytomegalovirus, can also be discovered without causing noticeable symptoms. Based on the presence or absence of lesions, the authors classified the detected CMV as a causative or uncertain pathogen.
Another significant result was that the treatment regimens for 86 out of 117 neonatal patients were altered based on mNGS data, with the majority of them (80/86) totally recovering and being discharged and 44 out of 86 patients completely or partially discontinuing the unneeded medication.
Limitations
- In this work, mNGS assays on DNA or RNA were done selectively. Though the majority of neonatal patients (158/168) recovered after assessment and treatment by doctors, insufficient detection may exclude causative or latent microorganisms, slightly prolong treatment duration, and raise the economic burden.
- Given the difficulty of collecting CSF samples from neonates, many more mNGS tests on neonates with CNS-bloodstream coinfection should be performed to determine whether mNGS testing of blood samples can replace that of CSF or respiratory tract samples.
- Data on the prognoses of neonates who were not diagnosed based on mNGS results should be collected to further understand the critical role of mNGS in clinical treatment.
Article Source: Reference paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Shwetha is a consulting scientific content writing intern at CBIRT. She has completed her Master’s in biotechnology at the Indian Institute of Technology, Hyderabad, with nearly two years of research experience in cellular biology and cell signaling. She is passionate about science communication, cancer biology, and everything that strikes her curiosity!