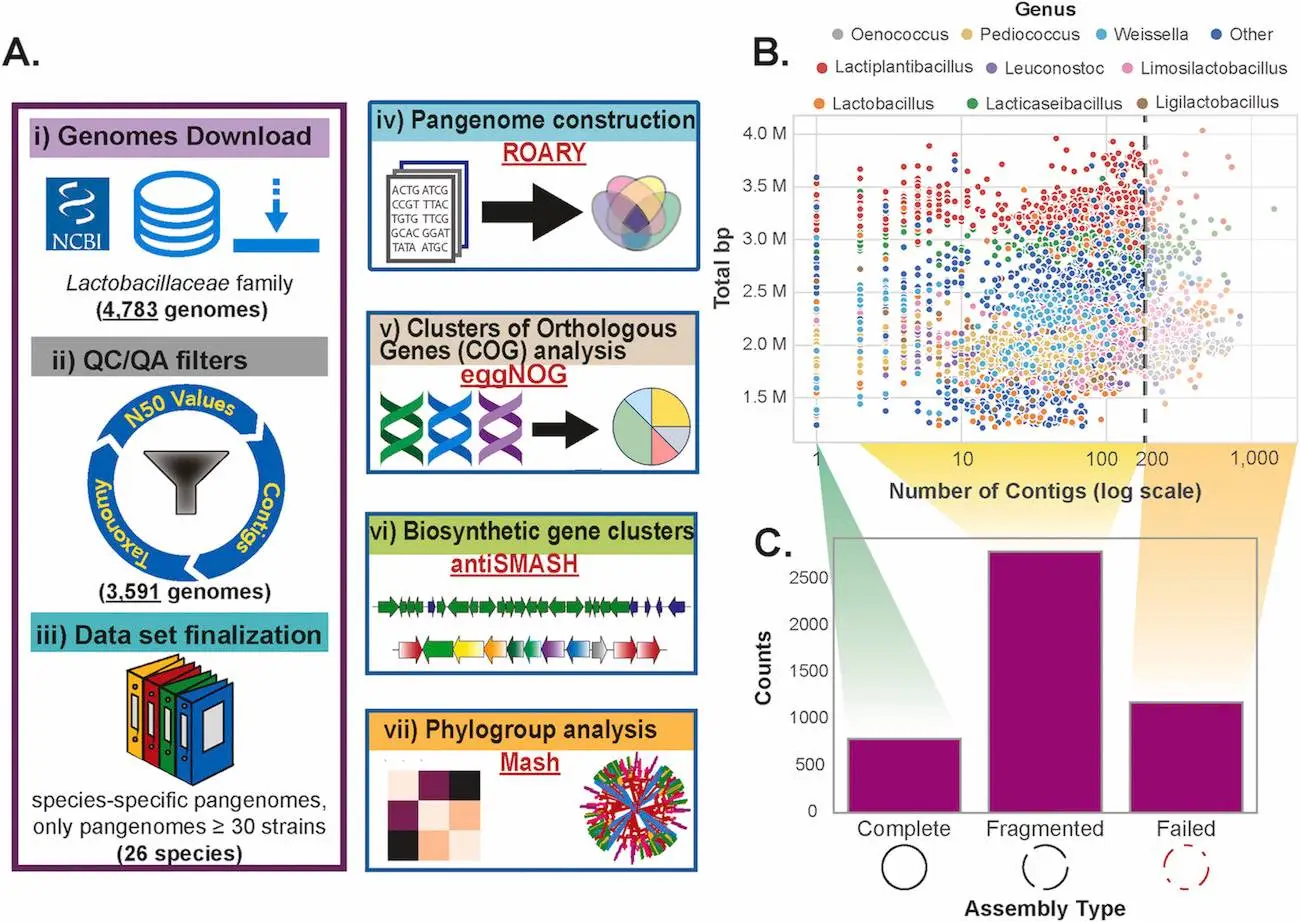
Researchers from the University of California, San Diego, and the Technical University of Denmark have conducted a comprehensive pangenomic analysis of the Lactobacillaceae family, a crucial group of microbes in the food industry. The researchers gathered 3591 high-quality genomes belonging to 26 species from publicly available sources and analyzed these genomes using computational tools. Key findings include the identification of core, accessory, and rare genomes, species-specific traits, and insights into biosynthetic gene clusters. This groundbreaking study offers the first comprehensive comparative pangenomics analysis of the Lactobacillaceae family.
A pan-genome can be defined as a whole set of genes that include all strains of the genus under study; basically, it is a fusion of all currently known genomes of the organism and its family. Lactobacillaceae contain a family of microbes that we are all too familiar with, and they are of immense importance to the food and beverage industries as well as to different ecosystems. This analysis found that the openness of the pangenome can correspond to the positions of transposons and mobilomes within the genome, as determined by observing their availability in the region. The researchers studied 26 species of the Lactobacillaceae family; among these, Lactiplantibacillus plantarum contained the largest number of genomes. The researchers also discovered groups of biosynthetic gene clusters with properties that can be useful for producing probiotics or for food preservation.
An introduction to Lactic acid bacteria
Lactic acid bacteria, or Lactobacilli, are friendly bacteria that are most popularly known for the benefits they provide us due to their presence in dairy products like milk and yogurt, as well as within the human body! They are important both commercially to humans and the natural ecosystem. They belong to the group of Gram-positive bacteria (those with thick cell walls) and come in two different types of shapes: cocci (spherical) and rod-shaped. They are different from other bacilli, given that they lack the ability to form spores for reproduction. Instead, they reproduce asexually through the process of binary fission, that is, the division into two individual bacterium containing copies of their parent DNA or RNA. On the basis of their ability to carry out the anaerobic fermentation of carbohydrates into lactates, they can be divided into two categories:
- Homofermenters: Lactic acid bacteria that only produce lactate as an end product.
- Heterofermenters: Lactic acid bacteria that produce ethanol and carbon dioxide in addition to lactate.
There exists a hypothesis that all lactic acid bacteria had a single common ancestor, and they later evolved to adapt to a soil environment. These evolutionary processes were accelerated due to the loss of some genes and through horizontal gene transfer, a process where genetic information is directly transmitted between organisms of the same genus. Members within the Lactobacillaceae family exhibit high genetic variability, most likely due to the differences in their lifestyles and adaptations to their own niches.
Importance of Lactic acid bacteria for humans
Because these tiny creatures have so many benefits associated with them, scientists and researchers have developed a keen interest in them. The ability of lactic acid bacteria to create specific conditions in order to protect their surroundings from infections and other dangerous microbes is one of their specialties. The following factors serve as the foundation for this ability:
- Metabolites: They are organic acids, such as lactic acid, that have antimicrobial and antifungal properties. They interact with their hosts, such as humans and plants, promote their health, and produce anti-inflammatory responses as well.
- Bioactive compounds: Bacteriocins based on peptides also contain antimicrobial and antifungal properties.
Both metabolites and bioactive compounds can be understood in depth with the help of publicly available lactic acid bacteria genomes.
- Competition: Lactic acid bacteria provide competition to undesirable organisms like fungi that can spoil food products. The bacteria has a rapid rate of carbon uptake and other essential micronutrients, which results in the gradual decline of fungi.
The first bacterial genome was successfully sequenced in 2001. Over the years, with the expansion of the Lactobacillaceae family, we can now perform comparative pan-genome analysis of different lactic acid bacteria species on a global scale.
Methodology for conducting pan-genome analysis
The following steps were carried out:
- Data was collected from the NCBI database and was subsequently annotated using the Genome Taxonomy Database Toolkit (GTDB-Tk). High-quality genomes were retrieved after performing quality control and quality insurance. The software Prokka was initially used for annotation.
- The pan-genome itself was constructed using roary software.
- Biosynthetic gene clusters (BGCs) were analyzed using antiSMASH software, as the detection of their presence can give the code for the synthesis of secondary metabolites. For comparing the BGCs, BiG-SCAPE was used.
- A phylogenetic tree is a representation of evolutionary relationships between organisms. In this study, it was constructed using autoMLST software. Another program, Mash, predicts the similarity between nucleotides belonging to two genomes and creates phylogenetic trees much faster than other pairwise alignment programs. It was integrated with in-house Python scripts to generate pairwise distances for all the genomes.
- Snakemake was used for most of the workflow.
Conclusion
The pan-genome analysis is important for helping us understand the evolutionary characteristics of lactic acid bacteria and their adaptability, both of which are caused by the diversity present in the bacteria’s genome. This study showcases that most species belonging to the Lactobacillaceae family have genomes that can be opened to a moderate extent. The size of rare genomes surpasses that of other genomes, indicating a much higher probability of discovering novel genes within them. The openness of a genome is an indicator of mobile genetic elements, enabling further research into horizontal gene transfer events. There is a need to fine-tune the resolution of the species that would be used for analysis. Rare genomes can be the key to finding novel strains that can be useful for food preservation and probiotic production.
Article source: Reference Paper
Learn More:
Swasti is a scientific writing intern at CBIRT with a passion for research and development. She is pursuing BTech in Biotechnology from Vellore Institute of Technology, Vellore. Her interests deeply lie in exploring the rapidly growing and integrated sectors of bioinformatics, cancer informatics, and computational biology, with a special emphasis on cancer biology and immunological studies. She aims to introduce and invest the readers of her articles to the exciting developments bioinformatics has to offer in biological research today.