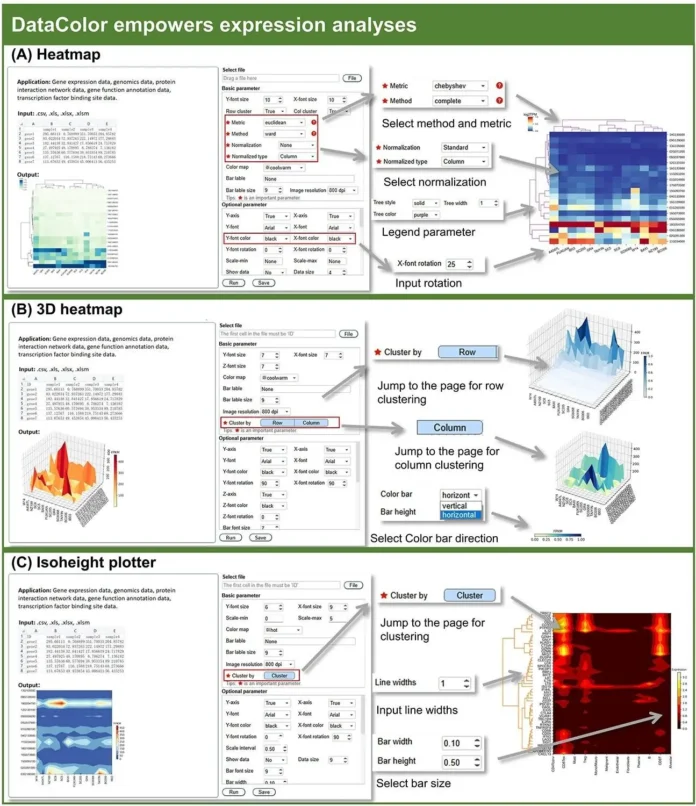
The display of many types of data with different orders of magnitude is a critical challenge in the rapid development of high-throughput omics technologies. Researchers from Hainan University and Sun Yat-Sen University present DataColor, a comprehensive software solution that has been painstakingly developed to address this issue and to close this gap.
DataColor is an all-inclusive toolbox made to manage various data types and deliver quick insights. With 23 different tools and more than 600 parameters, it uses the color spectrum to represent a range of data magnitudes and types. A wealth of insights concealed inside the complex relationships in the data are revealed by DataColor’s integration of sophisticated algorithms such as data clustering, normalization, squarified layouts, and adjustable parameters. A powerful tool for data research, this all-inclusive solution is perfect for traversing large datasets or displaying complex patterns.
Introduction
A rapidly expanding discipline, single-cell omics produces a variety of data from sources such as phenomics, metabolomics, and genetics. There are many difficulties in visualizing these data, particularly regarding magnitudes and patterns. Heat maps are a strong tool for data visualization, and they make it simple for users to identify data clusters and see how they are distributed. This has become more prevalent in genome assembly and gene expression investigations and allows for better-informed decision-making. Additionally, the electronic Fluorescent Pictograph (eFP) browser can be used as a visualization toolbox to examine the spatiotemporal expression of genes in individual organisms. However, there is still a shortage of comprehensive software made specifically for this purpose, which makes it difficult for researchers to evaluate and present their data efficiently.
Many bioinformatics software applications, pipelines, and packages for producing images, including heatmap tools, have been developed due to technological advancements. Though they still have certain drawbacks, these tools provide extensive visualization features beyond heatmap creation. For instance, they might not provide enough richness or parameters in the created heatmaps. Furthermore, it could be difficult for users to quickly and easily choose settings and debug heatmaps, and the final heatmaps might not always satisfy intricate specifications. Despite these obstacles, heatmap techniques continue to be useful tools for researchers who want to visualize complicated data sets. In the years to come, there is no question that improvements in technology and continuous innovations in heatmap tools will raise the caliber and accessibility of heatmap visualization.
DataColor: A One-stop Shop for Analyzing Different Types of Omics Data
The need for comprehensive, quick, simple, and intuitive information retrieval from big data is becoming increasingly important. To meet this demand, scientists created the DataColor tool, which uses the color spectrum to represent multiple data types and magnitudes. This tool may be used as a one-stop shop for analyzing different types of omics data for various applications. Researchers anticipate that large data access and visualization will never be easier or more effective thanks to DataColor.
DataColor is unique among analogous software in that it solely concentrates on using colors to represent a variety of data types and magnitudes. To uncover data connections and make it easier to integrate different types of massive biological data for study, it uses methods including data clustering, standardization, squaring, and parameter range adjustments. A broader range of users can utilize DataColor because it is made to be user-friendly and doesn’t require any programming skills, unlike other language libraries like ggplot2, D3.js, and Matplotlib.
Furthermore, DataColor offers several distinctive and cutting-edge tools. A wider variety of heatmap kinds and parameters are available with DataColor than with the web-based heatmap utility Heml. When it comes to the expansion of structural genomics applications, the development of 3D tools, the addition of background tools, and the comprehensiveness and variety of parameters, DataColor stands out from other software. It is beneficial when it comes to analyzing different facets of plant histology data. DataColor is expected to develop into a useful tool for analyzing and visualizing large biological data.
Limitation of DataColor
- Processing millions of data may be difficult if Python is used as the architecture.
- Interactive interfaces are now absent from DataColor; researchers intend to address this in further iterations.
Conclusion
DataColor is easy to use and requires no programming knowledge, making it accessible to a wider range of users. DataColor is a unique tool that uses colors to represent different types and magnitudes of biological data. It uses algorithms such as data clustering, normalization, squaring, and parameter range adjustments to reveal data correlations, making it easier to integrate different types of big biological data for research. DataColor can be used locally or as a software package for Windows, Mac, and Linux, meeting the needs of a wide range of users. A wide range of adjustable factors are available in DataColor, such as 22 distance metrics and seven clustering techniques. While the distance function gives a range of alternatives, these techniques offer a flexible examination of data relationships. DataColor’s heatmap color function uses cmap colors, which fall into four categories: Sequential, Diverging, Qualitative, and Miscellaneous. DataColor combines data analysis and visualization steps into a single workflow, significantly increasing operational convenience. Users can precisely depict the subtleties of data because of this extensive selection.
Article Source: Reference Paper | DataColor is available freely for all users on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.















