Scientists from the Broad Institute of MIT and Harvard have developed TEMPOmap, a method that unravels subcellular RNA profiles spatiotemporally at the single-cell level. The method incorporates pulse-chase metabolic labeling and multiplexed-three dimensional in situ sequencing for simultaneous profiling of the age and location of individual RNAs. The authors discovered kinetic gene clusters while performing RNA kinetic parameters across single cells. STARmap, which was also developed at the same lab at MIT for three-dimensional intact-tissue sequencing of single-cell transcriptomics, is the predecessor to TEMPOmap. The never-before capability of simultaneous spatiotemporal profiling of transcriptomics in single cells has now been achieved in TEMPOmap. This will enable the uncovering of molecular mechanisms underlying several biological processes in the future.
Spatiotemporal profiling on RNA: the past, the present, and the future
Spatiotemporal regulation of gene expression shapes cellular functions and states. The transcriptome’s heterogeneous expression in space and time is achieved through mRNA metabolism and trafficking. Spatiotemporal profiling of the transcriptome at the single-cell level is crucial for understanding transcriptional and posttranscriptional gene regulatory principles.
Currently, no method exists for simultaneous spatiotemporal transcriptome profiling at single-cell resolution. However, several previous methods have been developed for spatial transcriptomics at the single-cell level. STARmap provides transcriptional states of three-dimensional intact tissues. Other methods have also been developed for the spatial resolution of the transcriptome at the single-cell level. However, these methods can only produce spatial snapshots of cells and tissues but cannot determine the transcriptional dynamics due to the lack of a temporal component.
Separately, metabolic RNA labeling approaches such as scSLAM-seq have made it possible to profile a nascent single-cell transcriptome temporarily. Live-cell imaging techniques and approaches, as described in this review, have enabled tracking of the RNA trajectory inside cells. However, simultaneously visualizing multiplexed transcripts remains elusive.
Thus, a method for the simultaneous spatiotemporal resolution of the kinetic landscape of highly multiplexed transcripts at the single-cell level.
STARmap: the precursor to TEMPOmap
Spatially-resolved transcript amplicon readout mapping, STARmap, a platform for 3D in situ transcriptomics, was developed at the Wang lab at MIT. The platform integrates hydrogel-tissue chemistry, targeted signal amplification, and in situ sequencing. The method was able to retrieve gene expression information while retaining the 3D tissue anatomy. This was a remarkable advancement in method development for spatially resolved transcriptomics. STARmap is the predecessor to TEMPOmap.
TEMPOmap (Temporally resolved in situ sequencing and mapping): A brief overview of the method
The new in situ transcriptomic platform, TEMPOmap, enables simultaneous spatiotemporal profiling of RNAs at the single-cell level. The platform integrates metabolic labeling coupled with selective amplification of a pulse-labeled nascent transcriptome. The method further incorporates state-of-the-art 3D in situ RNA sequencing techniques with nanoscale resolution. The pulse labeling enables tracking of key kinetic parameters of the transcriptome. Thus, simultaneous profiling methodology is achieved.
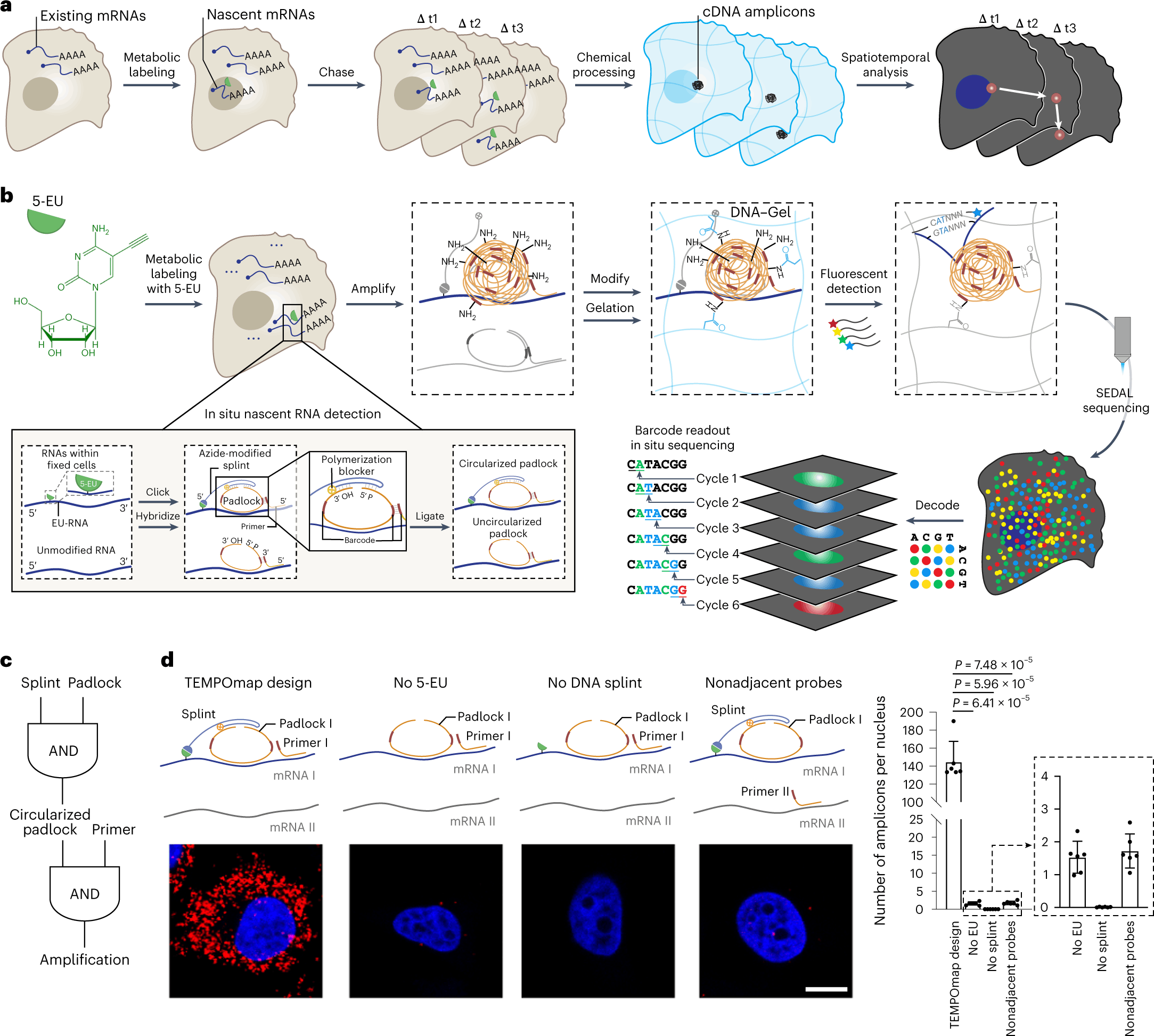
The following figure illustrates the TEMPOmap workflow.
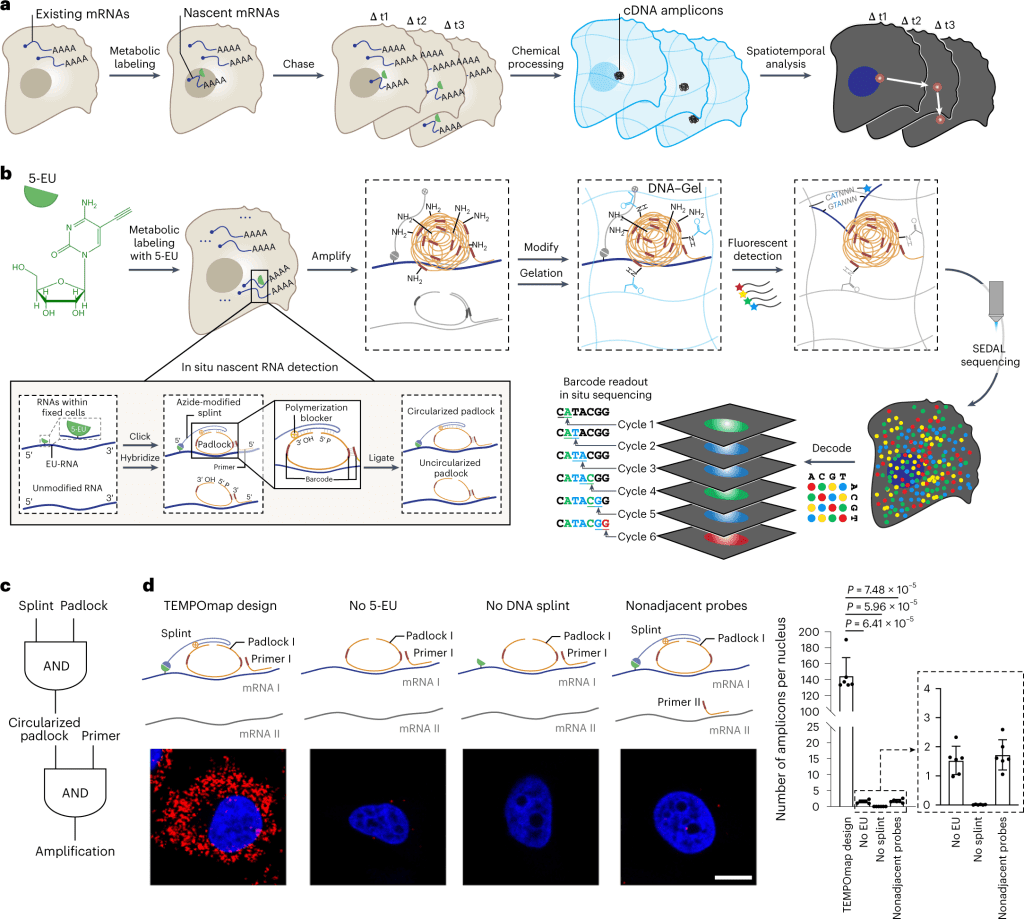
Image source: https://doi.org/10.1038/s41592-023-01829-8
The story of a lifetime: TEMPOmap reveals RNA dynamics from birth to death
TEMPOmap reveals the RNA dynamics over its entire lifespan, both temporally and spatially. TEMPOmap enables the resolution of the spatiotemporal evolution of single-cell transcriptome. It is also possible to determine the subcellular kinetic landscape of RNA across its lifespan. This allowed the authors to discover four gene clusters whose RNA temporal profiles coupled with cell-cycle phasing. The authors further probed to determine whether such a correspondence exists between RNA kinetics and global gene functions. This clustering analysis revealed five gene clusters with distinct kinetic landscapes, distinct single-cell expression, along with subcellular distributions over time.
The authors also applied TEMPOmap to study the spatiotemporal dynamics of mRNAs in cardiomyocytes and fibroblasts. This analysis revealed the kinetic landscape of several genes, nearing a thousand in number, and also revealed that mRNAs associated with genes with a specific function are synthesized faster and remain highly functional in particular cell types.
Conclusion
The spatiotemporal resolution of RNA dynamics had never been achieved before the development of TEMPOmap. Revealing the kinetic landscape of the transcripts now allows scientists to study the underlying mechanisms of several biological processes. The determination of the gene clusters revealed the correspondence between RNA kinetics and gene functions. This is indeed a remarkable and groundbreaking method, as it will now aid researchers in probing intricate questions concerning the dynamics of the transcriptome and gene expression regulation. This will also aid in a better understanding of trafficking mechanisms in subcellular contexts and help in the development of better drug delivery techniques. The authors envision that, with further optimization of labeling techniques coupled with several molecular probing approaches, TEMPOmap could, in the future, be adapted for in vivo and ex vivo tissue samples, thereby revealing the transcriptome dynamics therein.
Article Source: Reference Paper | Reference Article | TEMPOmap Availability: GitHub
Learn More:
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.