Researchers from the Children’s Hospital of Philadelphia, Philadelphia, USA, have created a versatile, easy-to-implement, and low-cost novel technique, TEQUILA-seq, for synthesizing large quantities of biotinylated capture oligos for any gene panel, that overcomes a substantial issue in Long-read RNA sequencing (RNA-seq). TEQUILA-seq’s ability to consistently and significantly enrich transcript coverage without interfering with transcript quantification while downsizing the expense of performing targeted sequencing of full-length transcripts and its exemplification of identifying novel transcript isoforms envisage its prospects in fostering future biomedical research.
Reducing Per-reaction Expense of Targeted RNA Sequencing with TEQUILA-seq
While genomic constituents typically remain similar across cells, their expression level dictates the whole cellular landscape. Quantifying the expression level of the genes is done by checking the level of transcripts, or more specifically, mRNA.
Notably, the large variations in our proteome levels are contributed by the modifications in the transcripts. Majorly, Alternative splicing and Alternative polyadenylation bring about the diversification of the gene products since these post-transcriptional processes render transcript isoforms.
This RNA-level regulatory process alters the further fate of the transcript regarding its translational products, localization, or stability. Identifying the transcript isoforms is key to a meticulous understanding of cellular and disorder phenomena.
RNA-seq is a powerful technique for obtaining ideas about the cellular transcriptomics profile, which reflects tremendous amounts of data regarding a cell. The issue here is that the most widely applied technology for exploring transcriptome profiles is short-read sequencing.
The short-read sequencers cannot read the entire transcript length, but it yields high throughput. When it is aimed to examine transcript processing events and transcript isoforms which exist in extraordinarily low concentrations, short-read sequencers cannot offer a credible output.
On the contrary, long-read sequencing techniques can supplement this research need since they can execute sequence full-length, from transcript’s one termini to another, but the throughput becomes much lower comparatively, and the expense increases undesirably.
Therefore, to meet the demand for a sequencing technique that will produce full-length sequence read as well as offer high coverage to confidently quantify transcript level and measure transcript isoform proportion along with providing the opportunity to discover rarer isoforms, the ‘targeted sequencing’ method was devised.
Targeted RNA sequencing enables accuracy and qualitative analysis and quantitation of the target of interest. Here, the transcript coverage can be enhanced by enriching the target followed by full-length sequencing.
One such efficient Targeted long-read RNA-seq is performed through Hybridization capture-based enrichment procedure where biotinylated capture oligos such as RNA Capture Long Seq (CLS) are used to target the desirable transcripts for enrichment selectively.
The major constraint here is the per-reaction cost of the biotinylated oligos, which again only can be reused for limited times. Philadelphia-based researchers solve this issue through their novel development of Transcript Enrichment and Quantification Utilizing Isothermally Linear-Amplified probes in conjunction with long-read sequencing, abbreviated as TEQUILA-seq.
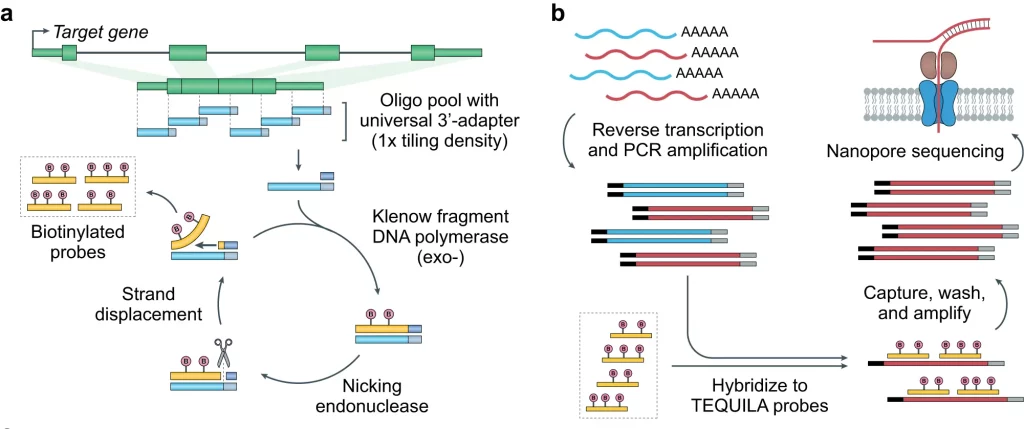
Image Source: https://doi.org/10.1038/s41467-023-40083-6
TEQUILA-seq: Features and Advantages
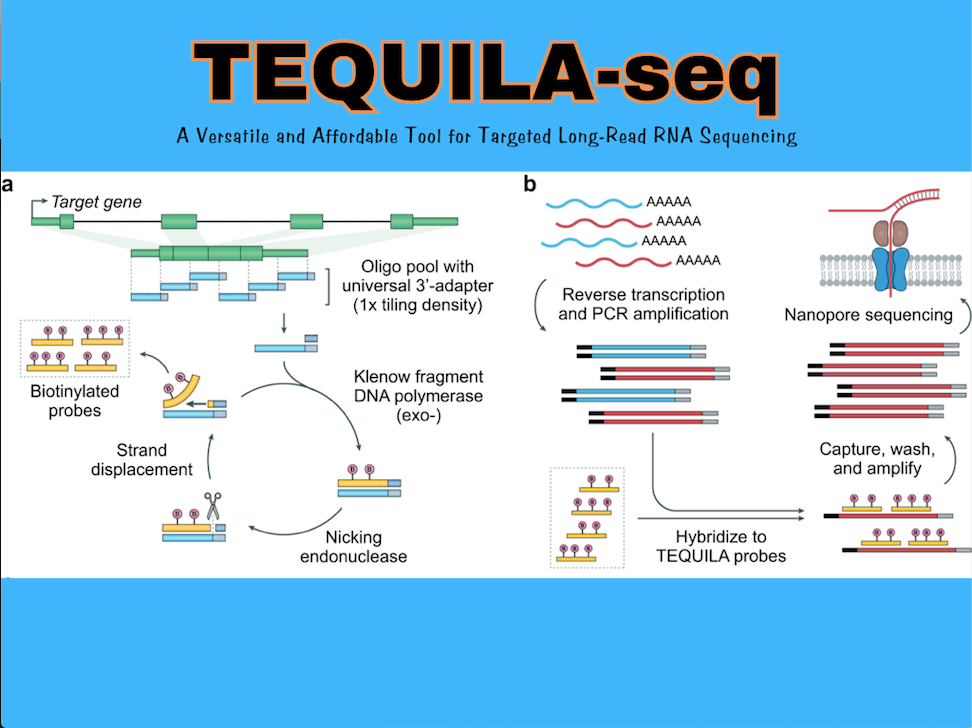
The enigma behind this cutting-edge technology lies in the concocted strategy for preparing capture oligos. The researchers formulated a nicking-endonuclease (nickase)-triggered isothermal strand displacement amplification (SDA) for synthesizing large quantities of biotinylated capture oligos from an array-synthesized pool of non-biotinylated oligo templates.
Single-stranded DNA (ssDNA) oligos are designed to tile across all annotated exons of target genes and are synthesized using an array-based DNA synthesis technology. Next, TEQUILA probes are amplified from ssDNA oligo templates in a single pool using nickase-triggered SDA with universal primers and biotindUTPs.
SDA facilitates isothermal amplification of internally biotinylated oligos via repeated cycles of nicking and extension reactions using a strand displacement DNA polymerase and pre-designed nickase-targeted nicking sites, thus allowing large quantities of capture oligos to be generated from starting templates.
The pool of TEQUILA probes can be used to capture full-length cDNA molecules of genes of interest. Strategizing this procedure, TEQUILA-seq aspires to offer the added advantage of Targeted sequencing, making it highly cost-effective and scalable for large gene panels and sample sizes; as compared to commercial approaches.
With the incredible application of a low-cost ssDNA oligo pool and the generated large probe synthesis output, TEQUILA decreases the costs associated with targeted capture. TEQUILA can be complemented with long-read RNA-seq for yielding high coverage of full-length transcripts, thus enabling the identification and quantitation of transcript isoforms.
Along with downsizing the expenses, TEQUILA-seq doesn’t compromise on the desired target enrichment with respect to commercially available methods. Even for long transcripts, TEQUILA-seq achieves comparable and effective performance and improves the detection of transcript isoforms of target genes while retaining the relative proportions of the transcript isoforms.
Conclusion
The biomedical applicability of TEQUILA-seq was also evaluated by profiling full-length transcript isoforms of 468 actionable cancer genes across a broad panel of 40 breast cancer cell lines. The experiment resulted in the identification of a novel transcript isoform in extensively studied cancer genes such as TP53. Apart from that, scientists discovered absurdity in transcript isoforms of Tumor Suppressing Genes (TSG), which can be inferred as the RNA-associated mechanism for TSG inactivation.
TEQUILA probes now emerge as an alternative for performing hybridization of cDNA synthesized from reverse transcription of the mRNAs for further enrichment and full-length sequencing. Moreover, it is compatible with both DNA and RNA sequencing. The convenient generation of large quantities of biotinylated capture oligos for any target panel at a low cost with high efficiency and feasible application in targeted sequencing makes it prospective for impacting biomedical research for novel discoveries.
Story Source: Reference Paper | TEQUILA-seq Availability: GitHub
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.