Scientists unleash the potential of digital pathology data by training computer-aided diagnosis models and making use of convolutional neural networks without human annotations. The cancer diagnostics model developed in the study for empowering digital pathology limits the human effort to a minimum.
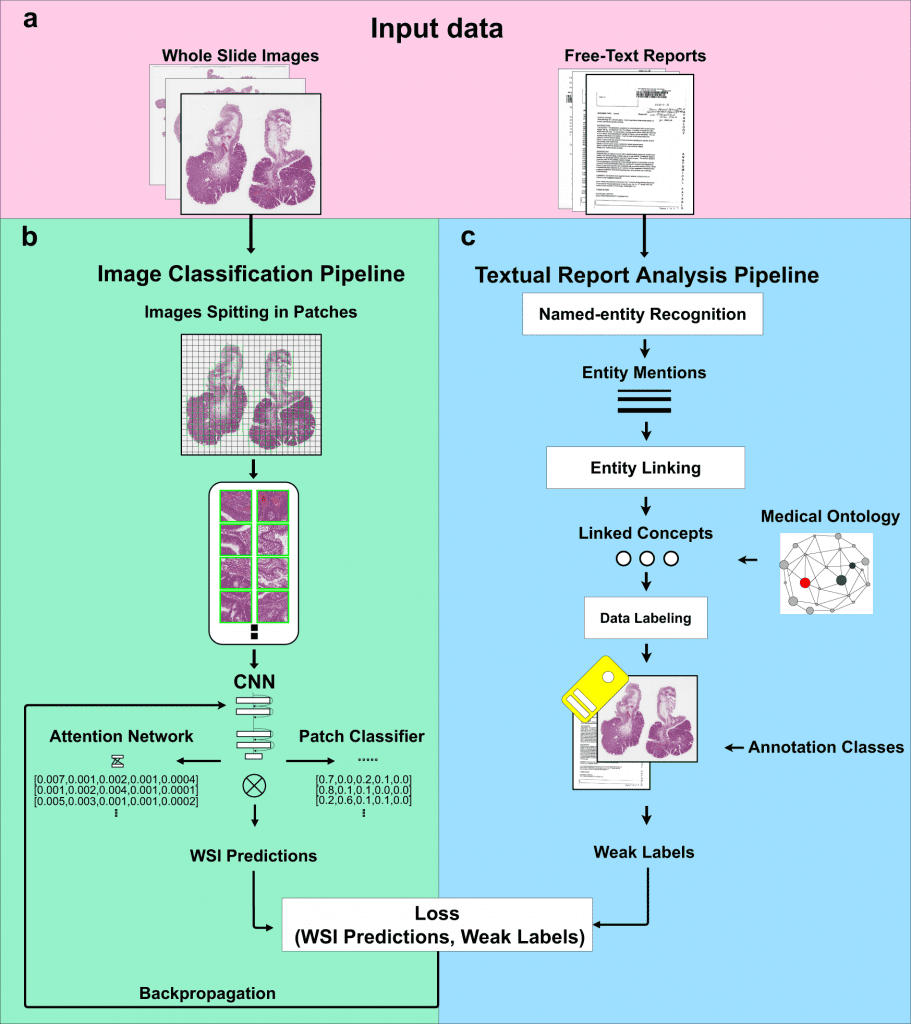
Image Source: https://doi.org/10.1038/s41746-022-00635-4
Deep learning algorithms’ improved performance and the digitization of healthcare operations are opening the door to novel approaches to cancer diagnosis.
However, the necessity for huge datasets to train reliable computer-aided diagnosis methods that can aim for the high diversity of clinical situations and data produced does not scale with the availability of medical practitioners to annotate digitized images and free-text diagnostic reports.
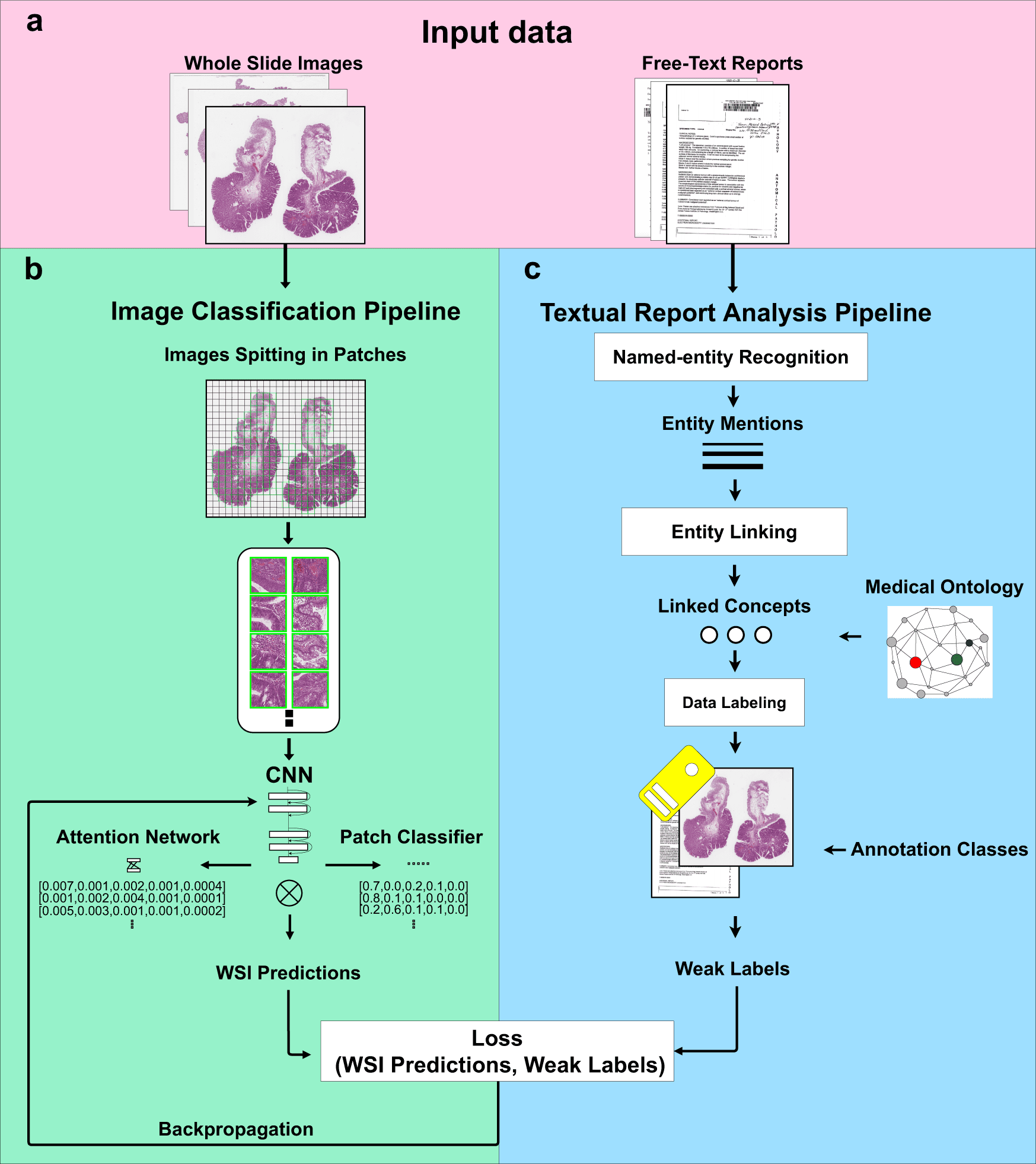
This study suggests and assesses a method for teaching computer-aided diagnosis tools about digital pathology without using manual annotations.
The method involves two steps: first, it uses diagnostic reports to automatically extract ideas with semantic significance, and second, it uses those concepts as weak labels to train convolutional neural networks (CNNs) for histopathological diagnosis.
The method is evaluated on more than 11’000 photos from private and publicly available datasets after being trained (by 10-fold cross-validation) on 3’769 clinical images and reports provided by two hospitals.
It is compared to the same architecture trained with manual labels and the CNN trained with automatically generated labels.
According to the findings, text analysis and end-to-end deep neural networks can be used to create computer-aided diagnosis tools that perform well at the micro-accuracy of 0.908 at the image level, using only pre-existing clinical data without the need for manual annotations.
Detailed Analysis and Subjective Evaluation of Findings
Although, still there are a number of obstacles to overcome, the digitization of clinical histopathology workflows and deep learning improvements are paving the way for Computer-Assisted Diagnostic (CAD) technologies that can learn from clinical data without human intervention.
The gold standard for cancer diagnosis is histopathology. It entails examining tissue samples to find disease symptoms at the microscopic level.
Biopsies or surgical resections are used to obtain tissue samples, which are then processed for microscopic analysis by a pathologist. The manual analysis takes up to an hour per image, making it time-consuming.
The inter-pathologist agreement on the diagnosis is typically low due to varied tissue morphologies, arbitrary selection of the tissue sections to study in-depth, and subjective evaluation of findings.
The rise in Numbers of Hospitals Digitizing WSIs
Even though digital pathology is becoming more widespread8, the processing and analysis are typically done in clinical practice with only a little amount of digital help. In digital pathology, entire slide images—digitized tissue specimens—are acquired and managed (WSI).
With a typical optical magnification of x20–x40, whole slide scanners capture images with a high spatial resolution of 0.25–0.5 m per pixel.
WSIs are typically saved in a multi-scale format, which enables pathologists to view various image details during analysis at various magnification levels.
A pathology case report often describes pathological findings, including discoveries through WSI analysis.
Semi-structured free-text reports are still the norm in clinical settings, despite the fact that synoptic reports (which include specific patient data in a structured format) are anticipated to become more popular in the future.
The type of tissue specimen, the findings discovered through examination, an early diagnosis, and the patient’s anamnesis are only a few of the fields that are included in semi-structured reports.
Hospitals are increasingly digitizing WSIs, enabling the collection of many images and diagnoses.
Heterogeneity in Stain Variations
A more modern field, computational pathology, uses technologies for computer-assisted diagnosis to automatically examine digital pathology pictures.
Convolutional neural networks (CNNs) have become the most advanced technique for completing a variety of computational pathology tasks with great performance.
The real potential of digital clinical pathology data hasn’t been released yet, and a number of problems remain, despite an increase in techniques, applications, and scientific discoveries.
First, in order to train models that can handle the high data variability of clinical practice, CNNs typically require huge datasets.
Second, fully supervised techniques, which offer the best results in computational pathology, call for pixel-wise annotations, which are difficult to acquire in medical settings because of their resource and time-intensive nature.
Third, WSIs are typically very large, making it difficult to manage and put them into memory, even with recent hardware.
As a result, it is customary and necessary to divide the WSIs into patches, however, doing so might occasionally introduce biases due to the loss of spatial linkages between the patches.
Finally, due to the lack of standardization in tissue preparation and acquisition between pictures and centers, WSIs can be highly diverse in stain variations.
Due to stain heterogeneity, models have difficulty generalizing to data from varied medical contexts, which may have stain variations that are different from those in the training data.
Weakly Supervised Learning Approaches
Weakly supervised learning techniques have been developed recently to address some of these problems.
Global (weak or image-level) annotations, as opposed to local (pixel-wise) annotations, are used in weakly supervised learning techniques.
Even though they are typically drawn from a specific and limited sub-region of the image, global annotations typically refer to the entire image.
For instance, even if the malignant tissue is present in only 1-2 percent of the overall image, a WSI would probably be classified as containing “cancer.”
In order to perform similarly to fully supervised techniques, weakly supervised CNN’s need training datasets that are larger.
On the other hand, global annotations offer the potentially game-changing benefit of being deduced from reports, which are frequently provided along with WSIs.
However, until recently, the majority of the time, weak labels from the report had to be extracted by medical specialists.
The Proposition in the Study
The difficulties that preclude fully utilizing digital clinical pathology for training-assisted diagnosis tools are discussed and evaluated in this research.
The suggested method combines a Natural Language Processing (NLP) pipeline for automatic free-text report analysis with a computer vision algorithm which is trained with sparse annotations for image classification.
When training an image classifier, the NLP pipeline automatically extracts ideas with semantic significance from free-text diagnosis reports.
Depending on the aspects of the problem to be solved and the state-of-the-art algorithm progress, the implementation of the method can be changed and adjusted, allowing for the adoption of various techniques.
The method is evaluated using data from digital pathology colons, entirely obviating the requirement for human intervention and maximizing the value of data gathered through clinical procedures.
The image classifier is contrasted with the same image classifier architecture that was trained using manual weak labels to show the reliability of automatically produced weak labels for training.
Training of the Proposed Approach
The Catania cohort and Radboud Medical University Center colon WSIs and reports are used to train the proposed approach.
It is then tested on both private and publicly available datasets using five classification classes.
The hospitals submitted the reports and the WSIs without any professional oversight or human data curation, making them a suitable test case for the suggested approach.
Colon is picked as a use case because of its significant social impact and its challenging diagnosis.
With an anticipated 75 percent growth by 2040 for both genders and a wide range of ages, colon cancer is currently the fourth most often diagnosed disease in the world.
The identification of malignant polyps, which are cell clusters that protrude from the colon’s surface and can belong to a number of classes, including adenocarcinoma, high-grade dysplasia (HGD), low-grade dysplasia (LGD), hyperplastic polyp, and normal, makes the diagnosis of colon cancer difficult.
The Endpoint
In future digital pathology, this study establishes a strong performance baseline for a methodology that can be used for the majority of diagnoses.
However, depending on the features of the problem to be solved and scientific developments, the components (such as the pre-processing algorithms and the data augmentation approach) implemented in this study can be altered and adjusted.
The authors of the article are currently working on duplicating the tests on additional tissue kinds and classes, where the framework is predicted to function similarly, despite the fact that the paper concentrates on the classification of five types of colon results.
The suggested method can also be used to categorize images from other medical fields, such as computed tomography or magnetic resonance imaging, and it can use other extraction and classification techniques.
Achieving excellent classification performance by fine-tuning was not one of the study’s main goals because it was more important to show the approach and analysis methods.
By using more complicated structures, methodologies, and ways to handle the stain-variability of the photos, as intended for future work, or by using more images or reports, performance can be improved.
In conclusion, the framework that has been given is an innovation in the field of digital pathology.
The framework paves the way for increasingly dependable computational pathology tools, with the crucial benefits of being efficient, generalizable, and capable of reducing to zero the human efforts to annotate extremely large amounts of data obtained during regular clinical procedures.
Article Source: Marini, N., Marchesin, S., Otálora, S. et al. Unleashing the potential of digital pathology data by training computer-aided diagnosis models without human annotations. npj Digit. Med. 5, 102 (2022). https://doi.org/10.1038/s41746-022-00635-4
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.