Scientists from Xi’an Jiaotong University propose the Histopathology Markup Language (HistoML), a representation language with a flexible syntax and extensible structure, along with a controlled vocabulary (Histopathology Ontology) to represent semantics in whole-slide images (WSI), to address this challenge using Semantic Web technologies (SW).
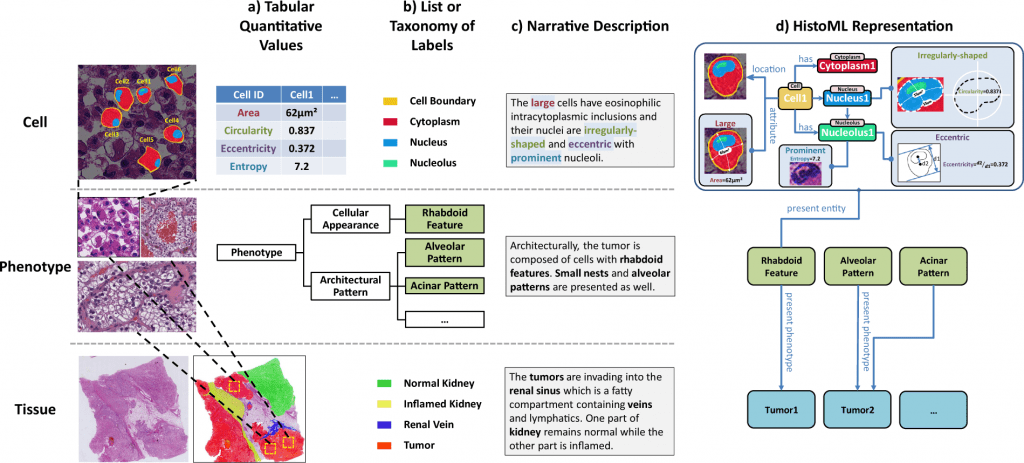
Image Source: https://doi.org/10.1038/s41597-022-01505-0
For cancer research and therapy, the study of histopathological phenotypes is essential since it connects biological pathways to disease prognosis.
To objectively characterize a histological phenotype often entails integrating heterogeneous histopathological characteristics in WSIs.
The fragmentation of histopathological features, which results from the absence of a standardized format and a controlled vocabulary for structured and clear expression of semantics in WSIs, has, however, hampered the widespread deployment of phenotypic characterization.
To close this gap, the scientists suggest the Histopathology Markup Language (HistoML), a representation of language and regulated vocabulary based on Semantic Web technologies.
HistoML might be used to express multiscale characteristics inside a WSI, from single-cell features to mesoscopic features, which is a crucial step towards the objective of making WSIs findable, accessible, interoperable, and reusable (FAIR).
To show the potential of HistoML-powered applications for phenotype characterization, the researchers pilot HistoML in representing WSIs of thyroid carcinoma and kidney cancer. They also use examples of HistoML representations in semantic queries.
Hindered Information due to Fragmentation
Cancer research and treatment heavily depend on the examination of histopathological phenotypes, yet it is still challenging to identify these phenotypes precisely because the spatial distribution of tumor cells is so highly variable and complex.
Large-scale extraction of histopathological features (such as cells, tissues, and phenotypes) from whole-slide images (WSI) is made possible by current advancements in digital pathology and deep learning approaches for image analysis, which also increase quantitative analytic techniques.
The combination of the numerous characteristics and the analysis’s findings shed light on how to describe histopathological phenotypes accurately.
For WSIs that adhere to the FAIR standards, it is lacking a uniform digital format and a tightly controlled vocabulary.
As a result, information fragmentation hampered the large-scale integrated study of histopathological phenotypes.
Elusive Intra-tumoral Spatial Heterogeneity
Histopathological phenotypes are the characteristics of tissues that a pathologist can see under a microscope after examining a biopsy or surgical material.
The variety of the individual components (such as cells, tissues, and substances) as well as their morphologies, spatial arrangements (such as architectural patterns), and behaviors all contribute to the complexity and variability of histopathological phenotypes (e.g., invasion, extension).
This has led to the realization that describing histopathological phenotypes requires an integrated examination of histopathological features.
Some researchers have successfully used this technology to characterize tumor-immune phenotypes because deep learning methods could automatically extract multiscale histopathological features from WSIs.
However, because of a paucity of FAIR WSI datasets, the widespread application of this technology for revealing intra-tumoral spatial heterogeneity has so far eluded researchers.
Accurate Representation within WSIs in Standardized and Machine-readable Format
As demonstrated by lymphoma and breast cancer, the overall volume of WSI data has entered a phase of rapid expansion.
Large amounts of information about histopathological phenotypes are also provided by a variety of deep learning models for histopathological image analysis, including segmentation, classification, detection, and quantitative analysis of multiscale histopathological features (e.g., tumor-level, tissue-level, phenotype-level, and single cell-level).
Unfortunately, the data is kept in several file formats, such as CSV, JSON, and XML, and is represented using unique representation techniques, which are widely used by hospital information systems, annotations of histopathology datasets, software tools, and computational models.
These representations lack a common controlled vocabulary and are rigid and ambiguous, creating a diverse collection of resources that are very challenging to combine and reuse.
A semantic standard is required to generate large-scale FAIR whole-slide images data to perform integrated analysis of histopathological phenotypes, but it is still difficult to precisely and completely capture the complex information contained within WSIs in a standardized and machine-readable format.
Exemplifying the Utilization of HistoML Representations in Semantic Queries
In this paper, the scientists propose the Histopathology Markup Language (HistoML), a representation language with a flexible syntax and extensible structure, along with a controlled vocabulary (Histopathology Ontology) to represent semantics in WSIs, as a means of addressing this challenge using Semantic Web technologies (SW).
System biology, integrative neuroscience, bio-pharmaceutics, and translational medicine are just a few of the scientific disciplines that have profited from the integration solutions made possible by SW.
While there are various undeveloped standards for highly multiplexed tissue photos, HistoML was established with a focus on histopathology images; in addition, HistoML is more advanced than the earlier works thanks to the technical advantages of SW.
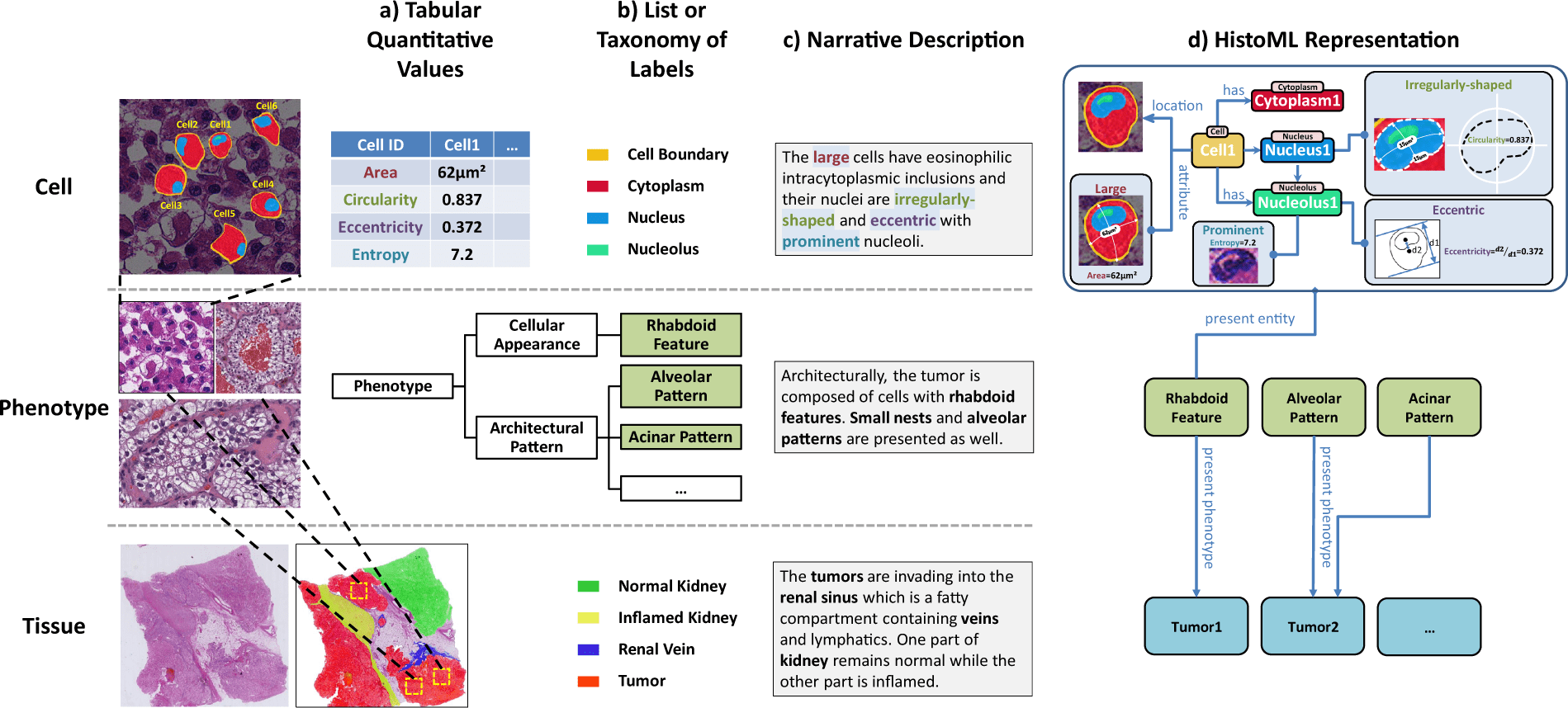
First, HistoML could represent mesoscopic-scale characteristics of histopathological phenotypes within WSIs in addition to single-cell features, such as the spatial arrangement of tissues, which is challenging to represent and consequently absent in many openly available atlases of human tissues and tumors.
Second, HistoML, which is a markup language based on Web Ontology Language (OWL), was able to integrate different histopathological features and analysis results of whole-slide images, fragmented in the previous representations, into a coherent representation, with their relationships to one another explicitly specified, providing a systems-level view of histopathological phenotypes.
Third, the scientists offer Histopathology Ontology, which makes extensive use of a variety of widely-used ontological resources pertinent to histopathology.
They test HistoML in representing semantics in whole-slide images of thyroid carcinoma and kidney cancer as a way to validate our research. To show the possibilities of HistoML-powered applications for phenotypic characterization, they also provide examples of how HistoML representations are used in semantic queries.
The Endpoint
Histopathology Markup Language representations cover various histopathological features in WSI data as well as the metadata. HistoML is a machine-readable format with a flexible syntax and extensible structure.
HistoML’s combination of these traits may enable several applications. First, the usage of HistoML and Histopathology Ontology as a common language and regulated vocabulary for whole-slide image data will lessen the number of translations necessary for information sharing between various sources.
Additionally, it makes it easier to integrate whole-slide image data by uniformly and thoroughly representing the various histopathological features extracted from WSIs by deep learning methods.
Consequently, it opens the door to creating a knowledge base of histopathology features in addition to the raw data repository (such as The Cancer Genome Atlas).
As some of the greatest FAIRness techniques for the life sciences, such as UniProt and BioModels, this feature repository would be a valuable resource for pathology.
HistoML representations of WSI data may help with an integrated analysis of histopathological phenotypes. First off, HistoML streamlines the implementation of multi-dimensional SPARQL searches of histopathological features for academics and pathologists.
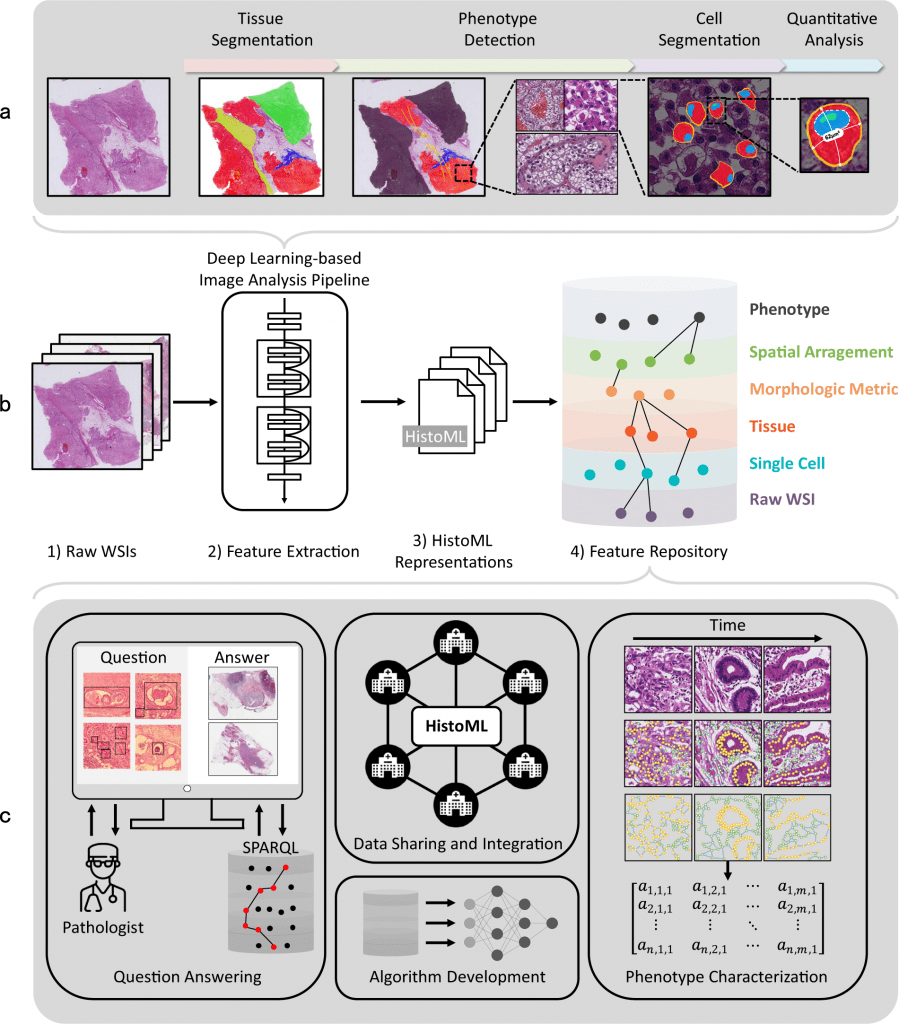
Image Source: https://doi.org/10.1038/s41597-022-01505-0
The feature repository could provide important insights into the intra-tumoral spatial heterogeneity by interacting with such a service.
However, this validation process, which is an essential step for accurate patient classification, typically takes years because the evidential cases are primarily gathered from medical publications and thus slowly accumulate.
For instance, it took ten years for acquired cystic RCC to be included in the WHO Classification of kidney tumors after its pathologically distinct features were first discovered. In comparison, this procedure would be significantly sped up by searching the feature repository.
The stacking of intricate HistoML representations of histological phenotypes and digital slides, on the other hand, offers a computational foundation for quantitative analysis of histopathological phenotypes.
To characterize the interactions between several scales of histopathological traits, many graph-based techniques, as well as additive models, could be applied to phenotype characterization, giving researchers new tools to define histopathological phenotypes systematically.
HistoML could aid in the computational study of whole-slide images in addition to phenotypic characterization. HistoML could enhance the performance and generality of deep learning models for whole-slide image analysis when used as a format for data annotation.
HistoML could annotate histological aspects within histopathology data more precisely and thoroughly than the present annotation formats for histology (such as list or taxonomy of labels and narrative descriptions).
As a result, it could provide datasets that encompass more variations that are seen in practical settings, leading to the development of more powerful image-processing algorithms to suit the changing requirements of pathologists and oncologists.
On the other hand, HistoML could be used as an information standard to unify various image-processing methods and WSI data types across research domains and programming languages.
This would allow for the construction of a FAIR computational pipeline for whole-slide image analysis. The pipelines in use now focus primarily on single-cell analytic techniques; HistoML would enable them to also include techniques for analyzing histopathological phenotypes.
The first step toward standardizing the depiction of all histopathological traits is HistoML Level 1. The demands of the community would be a major influence on how HistoML would develop. Further, the research community members look forward to promoting HistoML and using it in cancer research and treatment.
Article Source: Lou, P., Wang, C., Guo, R. et al. HistoML, a markup language for representation and exchange of histopathological features in pathology images. Sci Data 9, 387 (2022). https://doi.org/10.1038/s41597-022-01505-0
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.