Global environmental concerns and the need for sustainable development require new technologies for producing fine chemicals and managing waste efficiently. Nowadays, using enzymes in biocatalysis has proved to be more sustainable. Hence, identifying proteins with excellent biocatalytic properties from the vast array of possible sequences is a task that needs to be simplified. Jiahui Zhou and Meilan Huang from Queen’s University, Belfast, delve into the methodologies shaping the current landscape of enzyme design to provide insights into the state-of-the-art techniques and future directions in this field.
Introduction to Enzymes and their Design
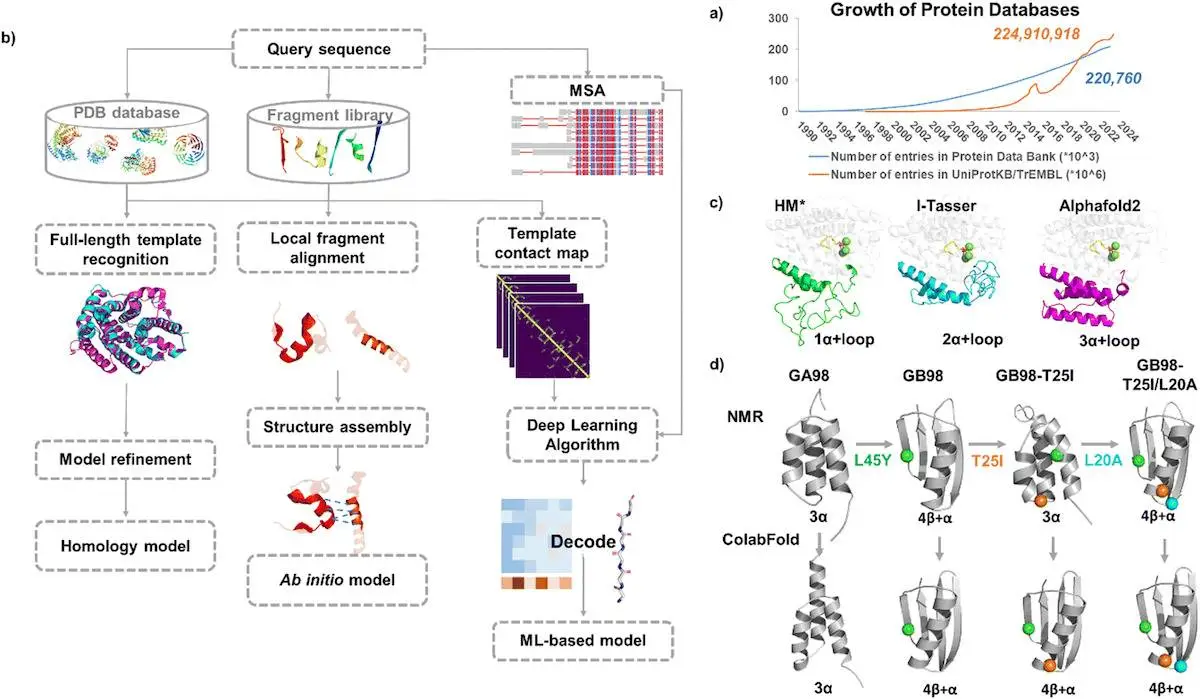
Enzymes are nature’s catalysts, facilitating biochemical reactions. The ability to design enzymes with tailored properties has a lot of potential to aid areas such as pharmaceuticals, biofuels, and synthetic biology. Computational methods can predict protein structures from sequences when 3D structures are unavailable. Prediction approaches include template-based modeling (homology modeling, protein threading) and template-free modeling (ab initio modeling). Tools like Modeller and Swiss-Model build homology models for sequences similar to known structures.
The review by Zhou and Huang provides a detailed exploration of the two main paradigms in enzyme design: molecular simulations and machine learning.
Molecular Simulations in Enzyme Design
These simulations offer atomic-level insights into enzyme dynamics, substrate interactions, and catalytic mechanisms. Zhou and Huang highlight several essential techniques within this domain:
Molecular Dynamics (MD) Simulations:
MD simulations allow researchers to observe the time-dependent behavior of enzyme-substrate complexes. These simulations are instrumental in identifying conformational changes and dynamic properties that are crucial for catalytic activity. GROMACS, AMBER, CHARMM54, and OpenMM3 are some of the MD software used.
Quantum Mechanics/Molecular Mechanics (QM/MM) Methods:
The QM/MM approach combines the minute accuracies of quantum mechanics for the active site with the efficiency of molecular mechanics for the surrounding environment. This hybrid method is particularly useful for studying reaction mechanisms and transition states.
Docking Studies:
Docking algorithms predict the preferred orientation of a substrate when it binds to an enzyme, facilitating the identification of potential catalytic residues and binding pockets.
Despite their power, molecular simulations are computationally intensive and require significant expertise to interpret complex datasets. The review emphasizes the need to integrate these simulations with experimental data to enhance their predictive accuracy.
Machine Learning in Enzyme Design
The advent of machine learning (ML) has introduced a transformative approach to enzyme design, enabling the analysis of vast datasets and the discovery of novel patterns. Such ML techniques discussed by the authors are mentioned below:
Supervised Learning:
Supervised learning algorithms are trained on a labeled dataset. Hence, these can predict enzyme properties such as activity, stability, and substrate specificity. Standard methods include decision trees, support vector machines, and neural networks.
Unsupervised Learning:
Unsupervised learning methods have no labeled data and have to find relationships within the data. Clustering and dimensionality reduction methods help identify hidden structures within enzyme data. These techniques are valuable for discovering new enzyme families and understanding evolutionary relationships.
Reinforcement Learning:
Reinforcement learning algorithms learn optimal strategies for enzyme design through trial and error with a reward system. This approach is particularly promising for optimizing enzyme sequences and catalytic efficiency.
Deep Learning:
Deep learning, with its multi-layered neural networks, has shown remarkable success in predicting enzyme functions and designing novel enzymes. Techniques such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs) are highlighted for their applications in enzyme classification and sequence generation.
ML models can be trained on simulation data to predict enzyme behavior more efficiently. The researchers underscore the importance of high-quality datasets and robust validation techniques to ensure the reliability of ML models.
Applications and Future Directions
The review by Zhou and Huang illustrates the practical applications of enzyme design in various fields:
Pharmaceuticals:
Designed enzymes may be used to develop new pills, improve drug delivery, and enhance healing efficacy. Enzyme-based totally biosensors and diagnostics are also rising as crucial tools in personalized medicine.
Biofuels:
Engineered enzymes break down biomass into fermentable sugars, producing biofuels, which are more sustainable than fossil fuels.
Environmental Remediation:
Enzymes designed to degrade pollutants and toxins can be employed in bioremediation efforts, contributing to environmental sustainability.
The authors also discuss future directions, emphasizing the need for interdisciplinary collaboration, advancements in computational power, and the development of more sophisticated models for enzyme design and prediction.
Conclusion
Zhou and Huang’s review provides a thorough and insightful overview of the current state and future prospects of enzyme design. The combination of molecular simulations and machine learning is shown to be extremely powerful for designing enzymes with unknown precision and functionality. As computational strategies continue to grow, the potential for innovation in enzyme design stays boundless in biotechnology and synthetic biology.
Article Source: Reference Paper
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!
















[…] Harnessing Computational Tools: How Machine Learning and Molecular Simulations are Transforming Enzy… […]