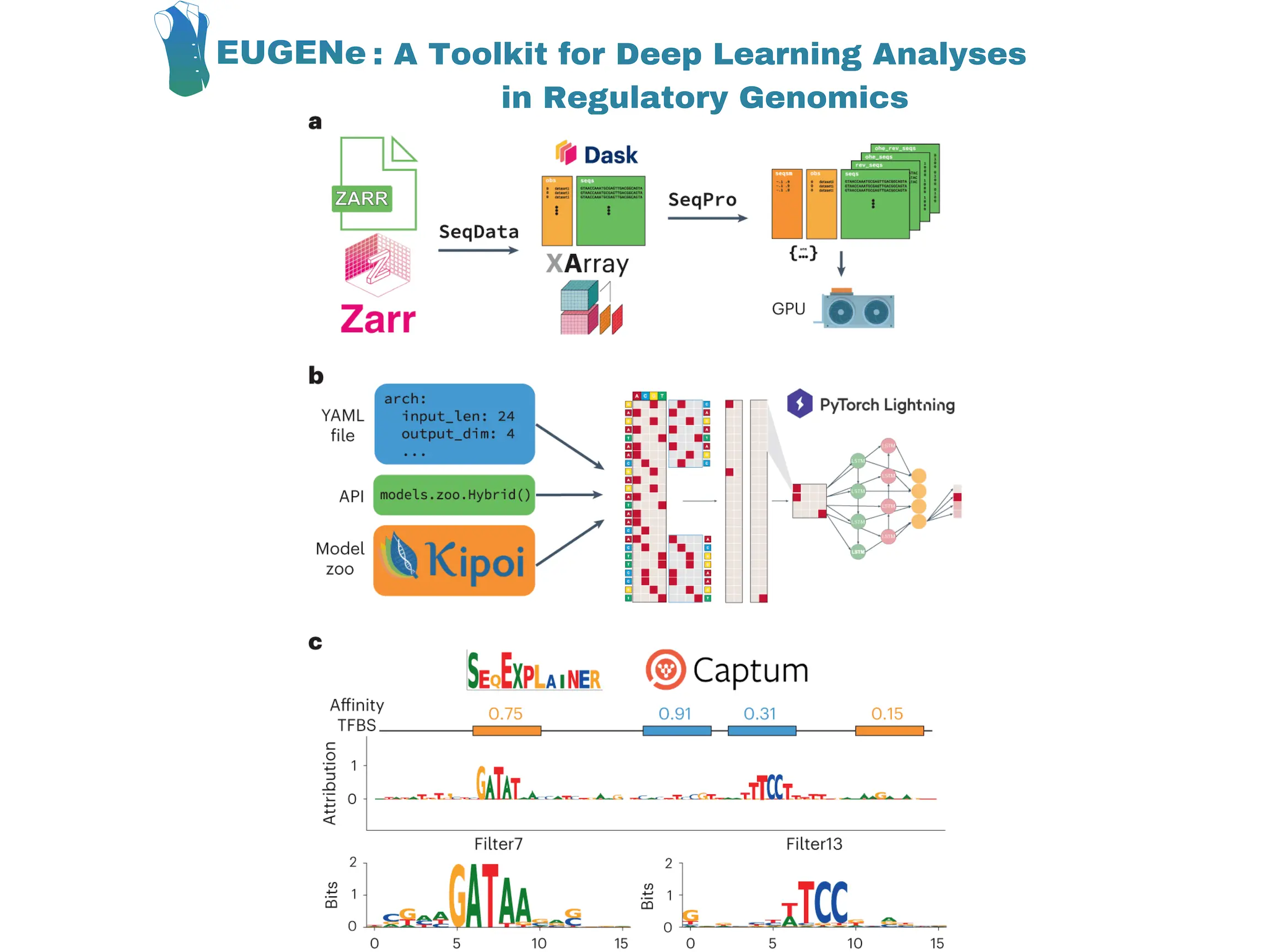
Researchers at the University of California, San Diego, built EUGENe (elucidating the utility of genomic elements with neural nets) – an end-to-end Python toolkit for analyzing genomic sequences using deep learning. EUGENe comprises a collection of modules and sub-packages designed to carry out the essential functions of genomics deep learning. EUGENe aims to enable computational scientists to swiftly enhance their understanding, create and exchange methods and models, and address significant questions about the genome and its functional encoding.
Understanding how genes are controlled by regulatory elements scattered across 98% of non-coding DNA represents a fundamental challenge in genomics. These mysterious regulatory sequences orchestrate the human genome’s precise spatial and temporal expression of over 20,000 genes. Cracking this “cis-regulatory code” could transform our ability to interpret disease risk variants in non-coding regions and program custom gene circuits.
While experiments like ENCODE have mapped biochemical activity across regulatory elements, making sense of these data remains enormously difficult. Machine learning, especially deep learning, has emerged as a promising approach to model sequence-function relationships and potentially reveal a grammar underlying the regulatory genome’s language.
However, effectively applying deep learning to problems in regulatory genomics has faced steep barriers to entry. Researchers at UCSD now aim to overcome these hurdles with EUGENe – an open-source Python toolkit to streamline regulatory genomics analyses using deep neural networks.
Why Deep Learning for Gene Regulation?
Various experiments profile the activity of regulatory elements by assaying proxies like transcription factor binding, histone modifications, chromatin accessibility, and spatial contacts. Deep learning leverages these rich data resources to infer sequence patterns driving regulatory outcomes.
Successes already demonstrated deep networks can:
- Discover binding motifs and predict transcription factor occupancy
- Forecast gene expression patterns from DNA sequence
- Learn chromatin structure and 3D genome topology behaviors
- Model massively parallel reporter assays quantifying regulatory potential
In addition to improving predictions, these models generate useful sequence representations to elucidate the grammar underlying regulatory functions. However, realizing the promise of deep learning in deciphering the regulatory genome has faced substantial roadblocks.
Hurdles in Applying Deep Learning to Genomics
While training complex neural networks has become simpler with libraries like PyTorch and Tensorflow, several genomics-specific difficulties persist:
Data heterogeneity – Diverse data types like sequences, arrays, signals, and graphs pose integration challenges
Complex workflows – Tasks need specialized splitting, architectures, and tuning lacking standardization
Limited tools – Existing packages have narrow functionality or arduous interfaces
Reinventing wheels – Labs duplicate effort instead of building on common frameworks
These impediments slow analytic iteration, harm reproducibility, and hinder methodological progress. There is a pressing need for findable, accessible, interoperable, and reusable (FAIR) deep learning tailored for regulatory genomics.
Introducing EUGENe – A FAIR Toolkit for Genomics Deep Learning
EUGENe’s architecture revolves around three key stages: extracting, transforming, and loading (ETL) data; instantiating, initializing, and training (IIT) neural network architectures; and evaluating and interpreting (EI) learned model behavior. The overarching goal of EUGENe is to offer a user-friendly interface for end-to-end deep-learning analyses, promoting effective design, implementation, validation, and interpretation of models in regulatory genomics.
To demonstrate EUGENe’s utility, the toolkit was applied to three distinct predictive modeling tasks, showcasing its flexibility and adaptability across different genomic datasets and tasks.
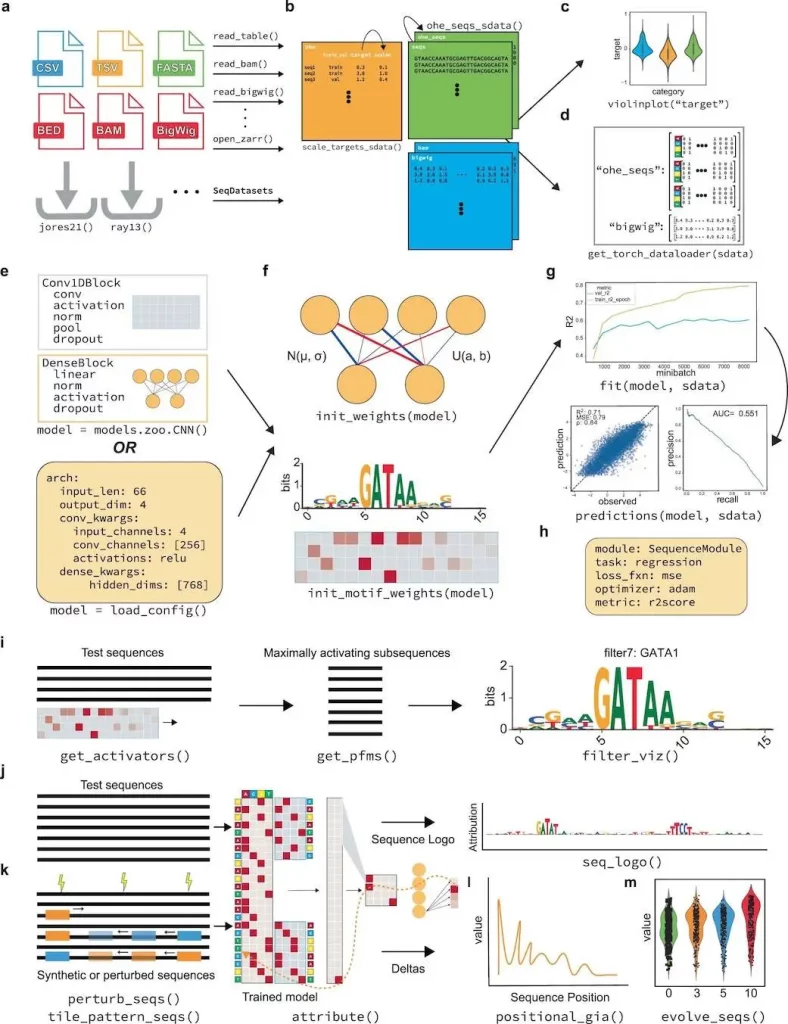
Image Source: https://doi.org/10.1038/s43588-023-00544-w
Case Study 1: Plant Promoter Activity Prediction
- Analyzed STARR-seq assay quantifying ~80K plant promoters
- Achieved performance matching bespoke classifiers
- Interpretation revealed core promoters like TATA-boxes strongly contribute
Case Study 2: RNA Binding Protein Specificity Prediction
- Implemented canonical DeepBind architecture and multitask networks
- Attained accuracy rivaling established tools like DeepBind itself
- Discovered models correctly learn binding motifs
Case Study 3: JunD Binding Classification
- Developed neural networks predicting JunD ( a transcription factor that binds to menin encoded by the MEN1 tumor suppressor gene) binding from ENCODE ChIP-seq
- CNNs significantly outperformed fully connected models
- The analysis highlighted meaningful sequence patterns linked to binding
Future Directions for EUGENe
While EUGENe presents a significant advancement in the genomics deep-learning toolkit landscape, the developers acknowledge opportunities for future development. The toolkit currently focuses on nucleotide sequence input and is poised to expand its functionality to handle protein sequences and multimodal inputs. As genomics assays transition to single-cell resolution, EUGENe aims to incorporate features that allow users to explore cell-type-specific regulatory syntax.
The researchers envision EUGENe as a collaborative ecosystem for deep-learning applications in genomics research. By fostering community development through tutorials, workshops, and a dedicated user group, they aim to address the evolving needs of the genomics research community.
Conclusion
EUGENe delivers an important step towards ecosystem-oriented, FAIR deep learning in regulatory genomics essential to unravel nature’s genomic regulatory code. Looking ahead, continued progress requires input from diverse stakeholders – from bench scientists to AI experts to ethicists.
If the genomics community can coalesce around common frameworks and priorities – while acknowledging open challenges like safety – the future looks bright for deciphering gene regulation, drastically furthering both basic knowledge and translational potential. EUGENe represents an important milestone in this collaborative journey to decode life’s software at the heart of our genomes’ non-coding majority – promising to transform how we combat complex diseases.
Story Source: Reference Paper | EUGENe is freely available under the MIT license on GitHub
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.