Proteins are incredible molecules that carry out nearly every function necessary for life. However, their staggering complexity has made designing new proteins an immensely difficult challenge. Now, AI is poised to change that. Scientists have developed Chroma, a groundbreaking AI system that can generate completely novel protein structures and sequences from scratch. This innovation could usher in a new era of protein engineering for biomedicine and materials science.
The Immense Potential of Synthetic Proteins
Naturally occurring proteins demonstrate tremendous diversity, with over 200,000 documented structures. However, researchers believe nature has only tapped a fraction of the possible protein space. Accessing more of this potential could enable transformative applications:
- New therapeutics like protein drugs tailored for specific diseases
- Sustainable biomanufacturing of fuels, chemicals, and materials
- Protein-based sensors, circuits, and computing devices
- Advanced materials like self-assembling protein matrices
However, manually designing functional proteins is enormously challenging. The space of possible sequences and structures is practically infinite. Iterative trial-and-error strategies struggle to navigate this vastness. A major innovation is needed.
Introducing Chroma – An AI for Protein Generation
To overcome these limitations, researchers at Anthropic and UCSF have developed Chroma, an AI system that can directly sample novel protein structures and sequences. Chroma is a generative model, meaning it learns a distribution representing plausible proteins. Key capabilities include:
- Generating 3D atomic protein structures from scratch
- Designing associated amino acid sequences predicted to fold into those structures
- Modeling full complexes, not just individual chains
- Scalable computation, enabling large structures
- Conditioning sampling on desired properties like shape and symmetry
Chroma opens up more of nature’s vast protein space for exploration and design.
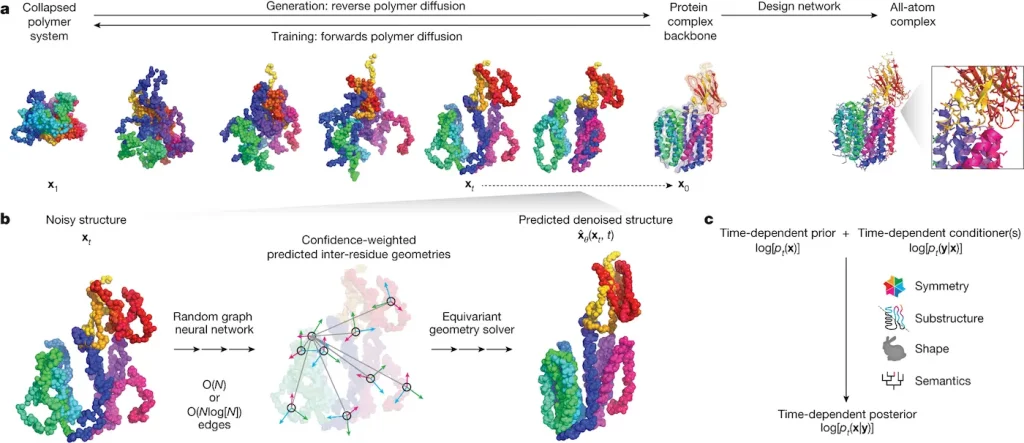
Image Source: https://doi.org/10.1038/s41586-023-06728-8
Sampling the Structural Diversity of Protein Space
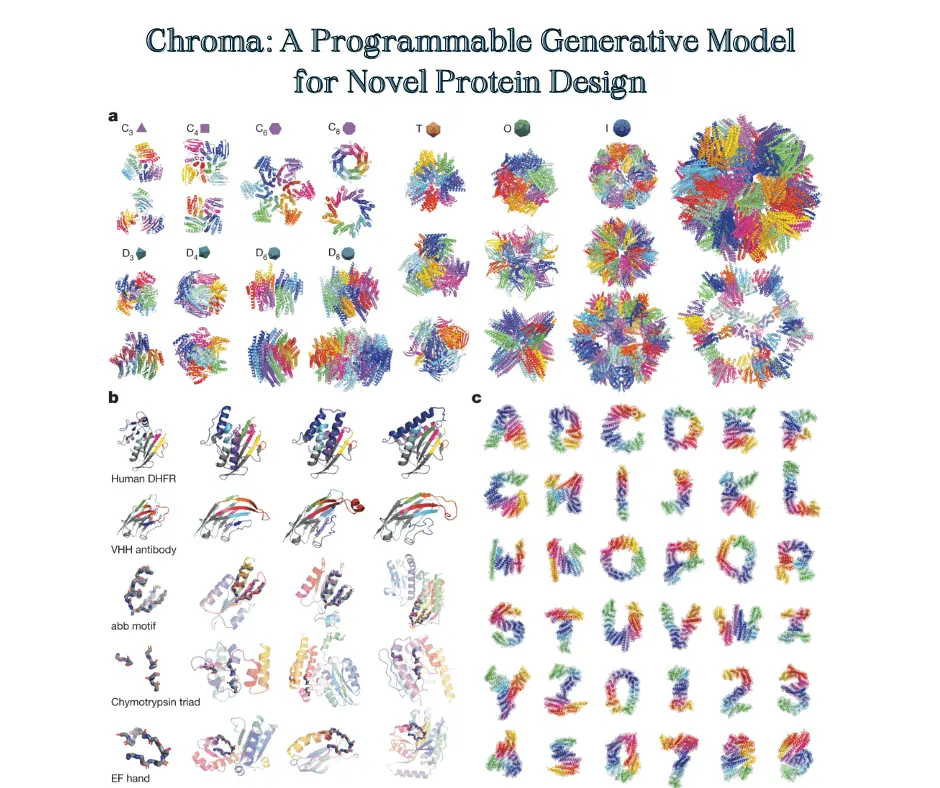
When generating unconditioned samples, Chroma produces remarkably diverse and complex structures reminiscent of real proteins. These include intricately bundled helices and sheets forming knot-free folds. Structural analyses reveal Chroma’s samples match natural protein properties like secondary structure ratios, compactness, and contact patterns.
Chroma also routinely generates structures with no close analogs in known proteins, especially for longer sequences. This highlights its ability to access unexplored regions of the protein structural universe.
Designing Sequences That Fold Into Intended Structures
However, generating backbone structures is only the first step. They must be paired with amino acid sequences that actually fold stably into those structures.
Chroma achieves this through a design network that generates optimized sequences given target backbones. In silico analyses show widespread agreement between generated structures and sequences. For many samples, independent protein folding algorithms confirm the designed sequences adopt the intended folds.
Early crystal structures of two Chroma proteins verify they fold precisely into the predicted conformations. This proves the structures coming out of Chroma are physically achievable.
Programmable Protein Generation with AI
But what makes Chroma truly revolutionary is its programmability. Users can steer the generative process towards desired properties without any retraining:
- Symmetry: Generate complexes with precise symmetric arrangements
- Shape: Conform proteins to specified 3D point clouds
- Language: Translate natural language descriptions into molecular structures
- Classes: Tailor structures to protein fold categories like alpha-helical or beta-barrel
These flexible constraints enable on-demand protein generation for function.
The Future of Computational Protein Design
Chroma demonstrates that generative AI can expand our ability to model and craft proteins. Looking ahead, Chroma could help design advanced protein materials, therapeutics, diagnostics, and more.
Challenges remain in extending functionality, ensuring safety, and integrating with biological testing pipelines. But by rendering protein design as an AI problem, solvable by data and computation, Chroma points to a future where proteins become as programmable as any other matter. The implications for biotechnology and medicine could be profound.
A New Era of Protein Programming is Dawning
Life has had billions of years to explore the protein design space. With tools like Chroma, AI and biological data may soon help us navigate what could be an infinite frontier of molecular possibility. Powerful techniques from deep learning are poised to revolutionize our ability to understand, manipulate, and build life’s most versatile molecules. The era of true protein programming may be rapidly approaching.
Article Source: Reference Paper | Chroma code is available on GitHub under the Apache 2.0 open-source license.
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.









[…] Enlightening the Proteomic Space with Chroma: A Revolutionary Generative Model for Protein Design […]