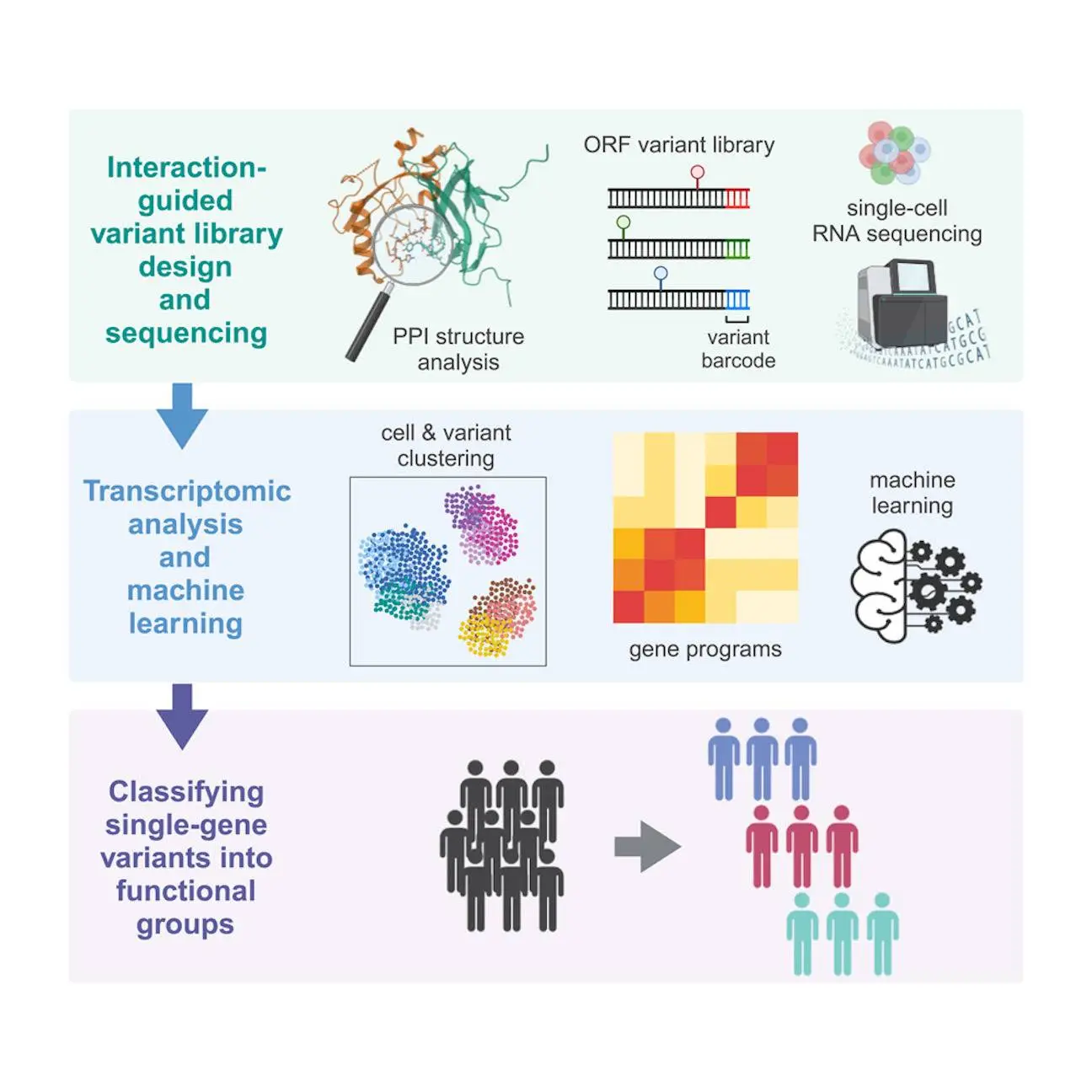
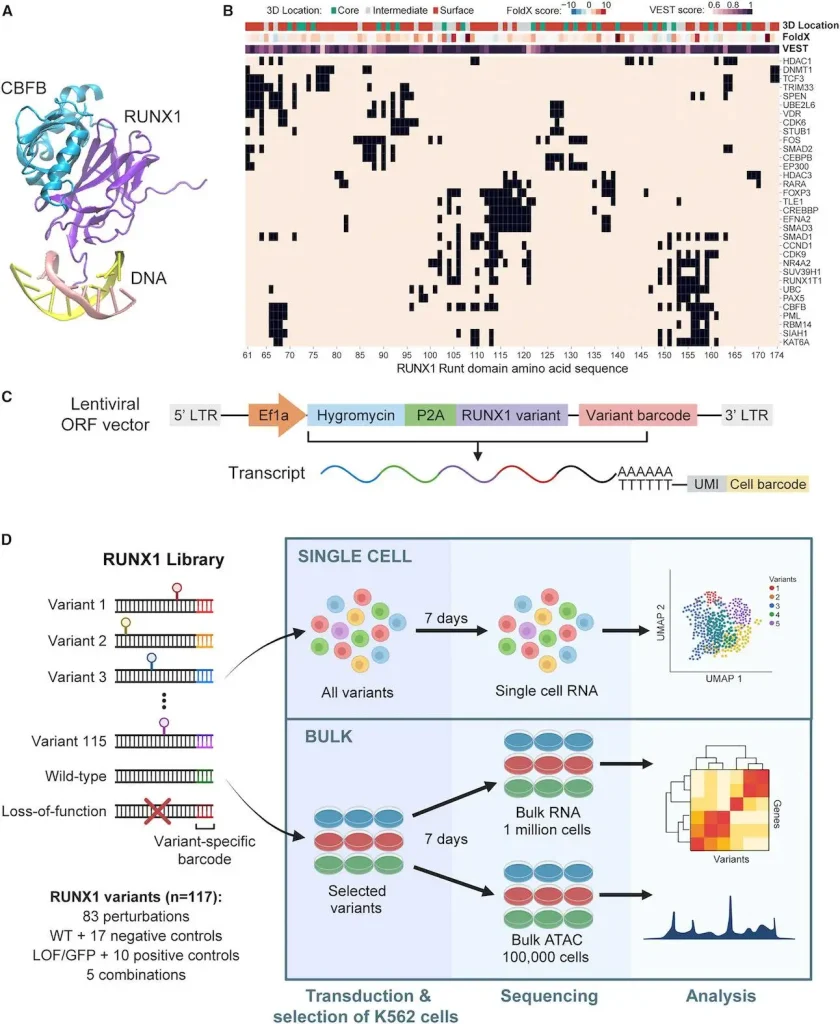
Researchers from the University of California deploy SEUSS (scalable functional screening by sequencing), a perturb-seq method, to analyze single-gene missense mutations, which have been notoriously difficult to interpret. The study focused on mutations in the RUNX1 gene, which is associated with myelogenous leukemia. Using SEUSS, the team evaluated 115 mutations and categorized them into three functional groups: wild-type-like, loss-of-function-like, and hypomorphic. This classification provides insights into how different mutations affect cellular processes.
The body of a human being is an intricate piece of engineering, and its functioning depends so much on genes that work in precise ways. Disturbance that can happen to such a delicately balanced system through gene mutations (alteration of the genetic code) may result in ailments including cancer. It is vital to comprehend how these mutations affect cellular processes, as this will enable the development of effective treatments.
RUNX1: The Maestro of Blood Cell Development
RUNX1 is a protein or transcription factor that binds DNA and regulates other genes’ expression levels. In the context of blood cell development, it has a critical function in different types, such as white and red blood cells. Leukemia is a type of cancer that starts in cells that make up blood-forming tissues, and people who have mutated forms of RUNX1 are more susceptible to this disease.
Addressing Situations on Protein Interface: An Innovative Way
Ozturk and his group concentrated on a certain kind of mutation known as protein-protein interaction interfaces. These are regions found on the protein’s surface interacting with other proteins. Some mutations in these locations may interfere with essential interactions, leading to an altered protein function.
The research team used Perturb-seq, a variation called SEUSS (Scalable Engineered Expansion of Short Unique Sequences). Using SEUSS, they were able to create and introduce a library of mutations at the RUNX1 Runt domain’s protein-protein interaction interfaces- which is quite important for DNA binding.
Research Findings by Leukemia Cell Examination
The team employed K562 leukemia cells as their model system. These K562 cells have alterations in the RUNX1 gene, making them suitable for studying additional RUNX1 mutations. Among these were:
- 83 variants of single amino acid substitutions at protein interaction interfaces,
- Wild-type (WT) and loss-of-function (LOF) controls,
- Negative control mutations (no effect predicted),
- Positive control mutations (known effect)
- Multiple interface mutation combinations
Revealing the Different Types of Mutations that are Functional
Gene expression profiles were analyzed in individual K562 cells with different variants to determine how these mutations affected the cells. From this analysis emerged three distinct functional classes of mutations:
Wild-type (WT)-like: These mutations had minimal effects on gene expression, indicating they did not significantly disrupt RUNX1 function.
Loss-of-function (LOF)-like: These mutations caused gene expression changes similar to those observed with complete RUNX1 loss, indicating a significant loss of function. Interestingly, a high proportion of LOF-like mutations clustered at the DNA binding site of the Runt domain, highlighting the critical role of this region for RUNX1 activity.
Hypomorphic: This class had a unique signature. While they shared some characteristics with LOF-like mutations, they also exhibited gene expression patterns suggestive of specific cellular responses, such as activation of pathways involved in neural growth factor signaling and neutrophil recruitment.
Chromatin Accessibility and Cell Fitness Beyond Gene Expression
Another area the researchers looked at with more detail was what happens when mutations occur by examining how tightly DNA is packed in the cell (chromatin accessibility). When they did this analysis, they found out that mutations that impacted RUNX1 function were associated with alterations of chromatin accessibility around genes normally regulated by RUNX1.
In addition, the team measured each variant’s abundance over time to know the impact of mutations on cell fitness. These helped to understand how K562 cells survived and proliferated due to these changes.
Further refining the Classification and Predicting Future Effects
The researchers didn’t leave it at that. They used their findings to update the classification of VUSs (variants of uncertain significance) and create a computer model for assessing the functional impact of new untested mutations.
The Intriguing Case of Hypomorphic Mutations
Hypomorphic mutations are particularly intriguing. However, this does not mean that these types of mutations only cause a partial loss of function similar to WILD-TYPE RUNX1. They also elicit specific cellular responses while representing a partial loss of function concerning wild-type RUNX1. In addition, with neural growth factor signaling and neutrophil recruitment pathways being activated, it could be speculated that RUNX1 activity could be rewired through these mutations in unexpected ways.
Unveiling Single-Cell Diversity Beyond Bulk Analysis
The use of single-cell RNA sequencing (scRNA-seq) is one of the strong points of this study. Bulk RNA-seq, on the other hand, studies gene expression in a cell population on average. Nevertheless, scRNA-seq can detect the gene expression profile in individual cells and expose concealed heterogeneity within a homogenous population.
In this study, it was essential to perform scRNA-seq analysis to identify three distinct functional classes of RUNX1 mutations (WT-like, LOF-like, and hypomorphic). Subtle differences between these groups may have been obscured by bulk RNA-seq, which might have led to incomplete comprehension of mutation effects.
The Road Ahead: Challenges and Opportunities
Like every other scientific inquiry, the research of Ozturk et al. leads to further explorations. These may include:
Confirmatory Validation: Even though different categories of mutations were identified in the study, other experiments must be carried out to prove these classes at the protein or cellular level. Thus, one might use protein-protein binding assays to understand how these mutations affect its interaction with other proteins based on RUNX1. Functional assays measuring how mutations impact specific cellular processes regulated by RUNX1 can also be employed.
Clinical Translation: The developed computational model predicting the functional impact of RUNX1 mutations has great promise for clinical use. However, successful translation of this model into a clinically useful tool requires validation with patient sample data. Moreover, ethical dilemmas concerning the utilization of these models in patient management should be considered.
Incorporating Additional Layers of Complexity: The proteins among which the study was concentrated on interaction; nonetheless, other extraneous factors such as RUNX1 post-translational modifications (PTMs) affect its functioning. PTMs are molecular variations that occur in proteins after they are produced. These changes can affect the activity, stability, and ability to interact with other protein molecules. In the future, it could be important to determine whether mutations modify PTMs interactively to modulate RUNX1 activity for disease development.
Conclusion
This is a big leap ahead in our understanding of how RUNX1 mutations cause leukemia in Ozturk et al.’s study. Notwithstanding, by studying protein-protein interaction interfaces and employing sophisticated methods like single-cell RNA sequencing, the investigators have uncovered some subtler impacts of these dysfunctions. This information suggests that improved diagnosis tools could be used in developing more personalized therapeutics and, ultimately, better outcomes for patients suffering from diseases related to RUNX1.
Join the Conversation!
Comprehending the impact of mutations on diseases is an intricate riddle, but as each study comes through, we get closer to the whole picture. What are your thoughts about this research?
Share your thoughts in the comments below! Let’s keep the conversation going and advance our understanding of health and disease together.
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.