In a recent scientific breakthrough, a team of researchers from NVIDIA and Innophore, USA, came up with a holistic dataset of the predicted protein folding for each member of the proteome of human beings (the complete set of molecules encoded by human genes), representing a great advancement in protein science.
Introduction
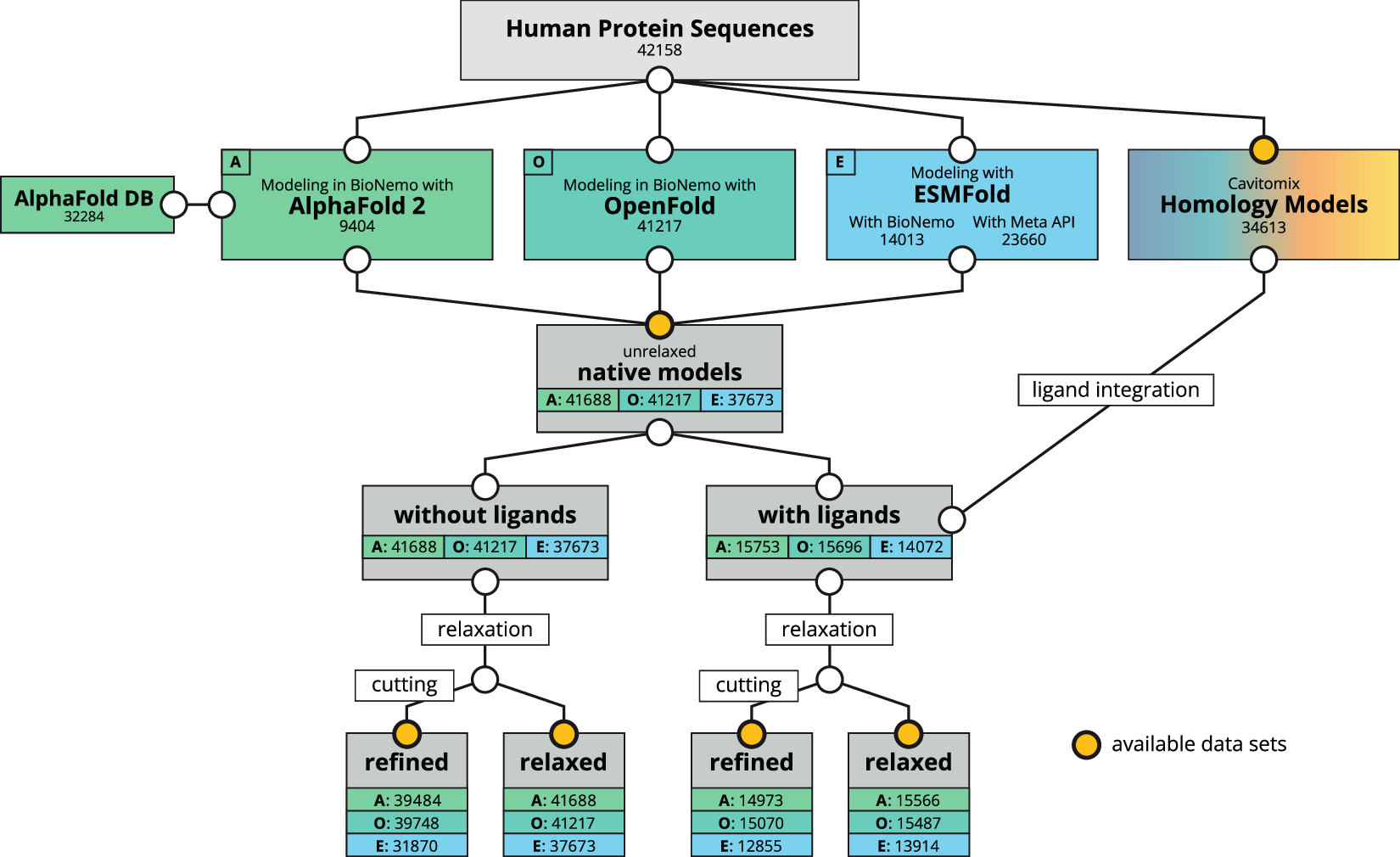
Life has protein as its foundation material, thus participating in almost every biological process. Medical advancements, drug innovations, and even the discovery of novel materials require understanding their complex designs, in other words, the structure of these proteins. However, protein structure determination is usually complex. In this study, scientists used modeling tools provided within NVIDIA’s BioNeMo platform and homology modeling platform from Innophore’s CavitomiX to create a reliable and accurate dataset.
Creating a Vast Dataset by Combining Protein Structure Predict Techniques
There are more than 20,000 proteins in the human proteome, each having a unique three-dimensional structure that governs its function. Protein structures were conventionally determined by X-ray diffraction or cryo-electron microscopy, which are time-consuming and demanding techniques.
This new dataset takes a different course of action. The researchers used advanced computational methods such as AlphaFold 2, OpenFold, and ESMFold to predict protein structures using artificial intelligence (AI) algorithms and powerful computers based on the analysis of protein sequences.
The dataset includes structures from different sources:
AI-predicted models: These are completely created by AI algorithms using amino acid sequence information for the proteins.
Homology models: These are structured based on similar sequences with known proteins.
By combining these techniques, the researchers were able to generate structural predictions for over 42,000 human proteins. This represents the most comprehensive collection of human protein structures available to date.
Revealing the Data: What is Inside the Dataset?
The dataset is full of treasures for researchers who want to learn about protein structure and function. It contains:
- Protein Information Includes protein details like UniProt accession codes, gene names, and amino acid sequences of each protein.
- Structural data: These are the most vital parts of the dataset as they contain predicted structures of proteins in PDB format, which is widely used to depict a protein’s structure.
- Quality parameters: Quality scores are provided to assess the reliability of prediction, such as pLDDT for AI-generated models and z-scores for homology models. The higher the score, the more confidence in the structure predicted.
- Several model versions: The dataset contains different variations to the models that include native (as predicted), relaxed (energy minimized), and refined models(with low confidence regions removed). In addition, there are other models that cater to potential small molecule binding sites (ligands).
Advantages and Uses: A Blessing in Disguise for Protein Science
In several ways, this dataset has tremendous potential to transform protein science:
Machine learning advances: The availability of a large number of protein structures can enable the development of robust machine learning models. Such models will be capable of learning from the data and provide even better predictions of protein structure with increased accuracy in the identification of functional relationships between proteins.
Drug discovery: Knowledge about protein structures is critical for designing drugs that target specific proteins involved in diseases. In this case, the dataset may be useful for scientists who develop new therapeutic agents.
Protein function prediction: With the help of protein structure analyses, one can understand how they interact with other molecules and perform their specific functions within a cell.
Material design: Protein structures may serve as guideposts for the development of new materials with peculiar properties. These biomimetic materials can find valuable resources in the dataset.
Tools and Resources Beyond the Dataset
Other avenues that can be explored by the data providers:
Additional modeling tools: The authors, however, know that there are more refined techniques like RoseTTAFold. Researchers are advised to consider such alternatives for generating even more diverse protein structure models.
Ligand integration with AlphaFill: Nevertheless, they advocate using AlphaFill as an alternative tool for ligand integration since it may lead to different results.
Refining models with AlphaCutter: The authors propose another model refinement tool known as Alphacutter, aimed at removing low-confidence regions from the models. It may have some useful insights for specific applications.
A Brighter Future for Protein Science: The Road Ahead
This comprehensive dataset is a significant step forward in protein science. By using AI and advanced computational methods, researchers now have a powerful tool for unmasking the secrets of proteins. Expect more sophisticated protein structure prediction methods to come up as AI continues evolving. Thereby, this will lead to the generation of more precise and various protein structure models.
Join the Conversation
Many scientific disciplines will find this research very interesting. However, what do you think about the ethical implications associated with using AI in protein science? How do you suppose someone can use this data set for medical research progress?
Please share your thoughts and queries in the comment section below! Let us talk about the future of protein science and how AI is likely to affect it.
Article Source: Reference Paper | The code used in the study is available on GitHub | CavitomiX plugin for Schrodinger’s PyMOL is available freely at https://innophore.com/cavitomix/ and https://pymolwiki.org/index.php/CavitOmiX
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.
















[…] BioNeMo Folds the Human Proteome: A Fused Dataset of 3D Protein Models for Machine Learning Applicat… […]