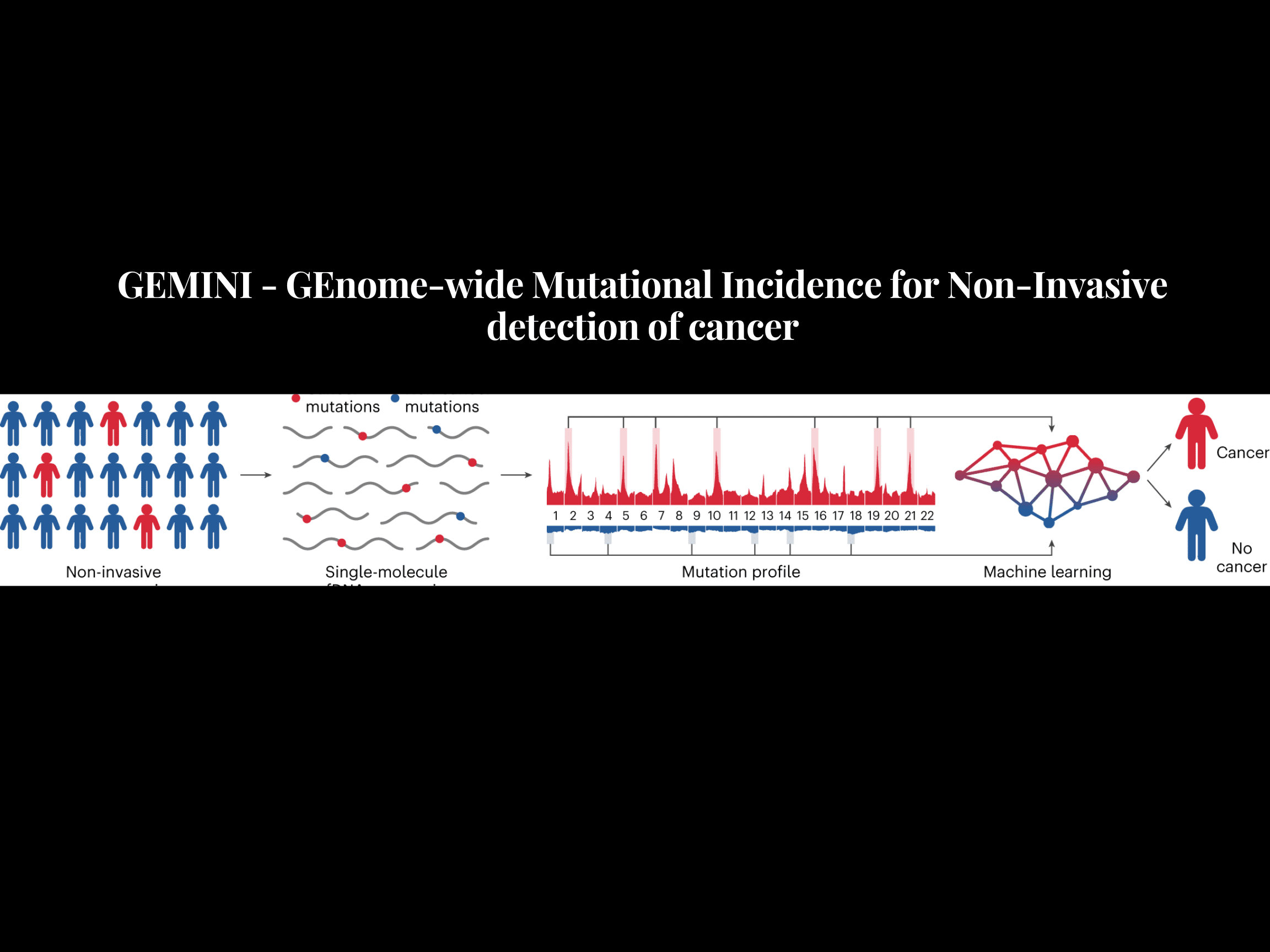
A new ray of hope for non-invasive cancer detection has emerged. A machine learning algorithm called GEMINI, created by John Hopkins University and Boston University researchers, can effectively diagnose cancer patients using CT scans, even in the early stages (stages I and II), without intrusive treatments like biopsies. This model focuses on the DNA changes in malignant cells that are present not just in tumors but also in the blood’s freely moving cells-free DNA (cfDNA). GEMINI successfully demonstrated accuracy in detecting lung cancer in more than 90% of the patients in the early stages itself.
The Main Problem in Early Cancer Detection
Many lives are lost to cancer primarily because of late detection, which also renders cancer treatment methods less effective. Early cancer detection certainly improves the chances of saving lives, but implementing effective screening methods is a huge hassle. A screening test called low-dose computed tomography (LDCT) is recommended for some high-risk individuals, but it comes with potential harm and low adherence rates.
Liquid biopsies offer a non-invasive approach to cancer detection by analyzing small fragments of DNA released into the bloodstream by cancer cells, called cell-free DNA (cfDNA). But, a hindrance to the efficacy of this method is the presence of minutely low proportions of cancer-specific mutations in cfDNA, which makes it difficult to distinguish those mutations from other background changes. The existing methods are confined to specific genes only, limiting successful early cancer detection.
GEMINI: Putting an End to Late Cancer Detection with Machine Learning
The research team, led by Victor E. Velculescu, hypothesized that analyzing the entire genome for cancer-specific mutations could be key to identifying increased cfDNA mutations related to cancer. Early detection of cancer could be accelerated if mutations in cfDNA can be recognized without any knowledge of the mutations in the tumor. And for this, it is highly essential that cancer-specific mutations in cfDNA are thoroughly distinguished from non-cancer-related changes.
They developed a machine-learning model named GEnome-wide Mutational Incidence for Non-Invasive detection of cancer (GEMINI) to detect cancer-specific mutations in cfDNA with great accuracy. They examined the DNA samples of 2,511 patients with 25 different cancer types and discovered that smokers had an average of 52,000 distinct mutations per genome.
Their goal was to identify cancer-related mutations in cfDNA even when it contained other background changes, hence they focused their attention on investigating the frequency of single molecule mutations in cfDNA. They emphasized a particular type of mutation, C>A, which is prevalent in cancers caused by smoking.
The researchers analyzed individual cfDNA molecules by reading their genetic information, and then they divided the genome into different sections (bins) of varying sizes. GEMINI then compared the mutation patterns and frequencies in the sites usually altered in cancer with those usually altered in normal cfDNA. This comparison allows probable cancer-related mutations to be enriched in the cfDNA, allowing the accurate differentiation between cancer-specific mutations and normal genetic variations found in healthy people.
Testing the Effectiveness of GEMINI
To test the effectiveness of GEMINI, it was made to analyze cfDNA from individuals participating in a lung cancer diagnostic trial (LUCAS), most of which happened to be at high risk for lung cancer due to their smoking history. It was found that certain regions in the genome had high occurrences of mutations in the tumor tissues as well as the cfDNA belonging to individuals with lung cancer, melanoma, and B-cell non-Hodgkin lymphoma, and these regions were linked with late replication timing.
Particular types of cancers were associated with particular types of mutations, such as lung cancer was associated with C>A mutations, melanoma with C>T mutations, and lymphoma with T>G mutations. It was also revealed that mutation patterns across tumors were consistent with those across cfDNA, indicating that GEMINI was capable of accurately detecting cancer-related mutations in the bloodstream.
The following inferences were made from the evaluation:
- GEMINI could accurately identify differences in mutation frequencies in individuals with lung cancer and those without, in the cases of both tumor tissues and cfDNA.
- GEMINI was successful in accurately detecting C>A mutations that are commonly associated with lung cancer.
- To measure the likelihood of lung cancer, GEMINI scores were used, and they were found to be higher in the individuals actually having lung cancer than the ones not having lung cancer.
- GEMINI exhibited great accuracy in detecting cancer mutations in multiple stages as well as subtypes of lung cancer.
- GEMINI was employed to analyze the cfDNA of seven individuals whose blood tests did not detect cancer but were later diagnosed with cancer. GEMINI was able to detect abnormalities in cfDNA mutational profiles, showing the potential for early cancer diagnosis.
GEMINI and DELFI: Enhancing Cancer Detection Using the Best of Both Worlds
DELFI is another method that employs cfDNA features. And the researchers wanted to determine whether combining DELFI and GEMINI could further improve early-stage lung cancer detection. Here is what they observed:
- Combining GEMINI and DELFI reduced the number of false negatives by 56% while maintaining a specificity of 80% and sensitivity of 91%.
- The combined approach showed an overall performance of 87% in early-stage lung cancer detection, outperforming both the individual methods.
- The concerns about missing cancers with this combined approach were reduced when individuals with lower GEMINI-DELFI scores received better prognoses, meaning their cancer was less aggressive.
Validating the Abilities of GEMINI
To validate GEMINI and GEMINI-DELFI, the researchers tested them on a validation cohort consisting of six high-risk individuals, mostly with stage-I cancer and others without cancer. The results are listed below:
- GEMINI scores were higher in individuals with lung cancer than those without lung cancer.
- The scores were even higher for people with advanced stages (stages III and IV) of cancer.
- The accuracy of GEMINI in the validation cohort was 81%, and it increased to 86% when it was combined with DELFI.
These results validate the cancer-detecting abilities of GEMINI as well as GEMINI-DELFI. Apart from detecting lung cancer with accuracy, GEMINI has been found to be effective in detecting other forms of cancer as well, such as liver cancer. GEMINI was also capable of monitoring the progress of cancer patients during treatment with GEMINI scores, as patients whose conditions improved showed lower GEMINI scores, and the GEMINI scores increased again in case cancer progressed despite the treatment.
Conclusion
Cancer is a fatal disease, and the most effective strategy to put an end to it is to detect it early before it spreads and conquers the major parts of our bodies. The existing methods for cancer detection encounter many hindrances to successful early cancer detection. GEMINI has been developed with the main purpose of detecting cancer in the early stages. GEMINI scores are an accurate measure of the likelihood of cancer happening to an individual, which can aid in subjecting the patient to required measures to prevent cancer from spreading, thus, increasing the patient’s chances of survival. GEMINI is effective across different forms, stages as well as subtypes of cancers. It shows high performance, which gets elevated further when DELFI is combined. This points to an optimistic direction in cancer detection as well as prevention.
Story Source: Reference Paper | GEMINI Code availability: GitHub
Learn More:
Neegar is a consulting scientific content writing intern at CBIRT. She's a final-year student pursuing a B.Tech in Biotechnology at Odisha University of Technology and Research. Neegar's enthusiasm is sparked by the dynamic and interdisciplinary aspects of bioinformatics. She possesses a remarkable ability to elucidate intricate concepts using accessible language. Consequently, she aspires to amalgamate her proficiency in bioinformatics with her passion for writing, aiming to convey pioneering breakthroughs and innovations in the field of bioinformatics in a comprehensible manner to a wide audience.