Researchers at the University of Missouri upgraded the MULocDeep web application by integrating animal, fungi, and plant species-specific models that meet competitive performance at the subcellular level against contemporary state-of-the-art methods and thus hold promising prospects in speeding up research in various domains, including drug development. Harnessing the power of Artificial Intelligence MULocDeep covers comprehensive datasets of 10 subcellular & 44 suborganellar localizations while quantifying the contributions of amino acid sequences of proteins or groups of proteins at single residue resolution and provides visualization featuring targeting mechanism analysis.
Understanding Protein Localization: Key to Cellular Activities
The physicochemical microenvironment around proteins is important for executing their destined functional roles in a cell. So After (Post-translation) or during (Co-translation), Proteins are transferred to cellular compartments or secreted outside the cell to exhibit their activities. Proteins inherently possess structure or sequence information or signals that indicate their target site. These molecular ‘postal codes’ are present at the terminal or internal amino acid sequences, which are recognized by sorting machinery that directs proteins to their respective organelle or compartments. For instance, proteins with nuclear addresses have the Nuclear Localization Signal (NLS), a short stretch of specific sequence that is recognized by shuttling systems to import the protein inside the nucleus.
Protein mislocalization paves the path to complicated diseases like cancer, metabolic disorders, schizophrenia, neurodegenerative disorders, epilepsy, and many more. Comprehending protein placement and the related machinery is useful for efficient drug delivery. For instance, Gamitrinib is a potential candidate for antitumor drugs since, owing to an amide-containing linker, it can be targeted to mitochondria and inhibit HSP90 in tumor cells.
Computational Tools for Predicting Protein Localization
Characterizing protein location in the wet lab is a laborious and expensive process where proteins are tagged with fluorophores [e.g., Green Fluorescent Protein (GFP) tags] and imaged using complicated microscopy techniques. But opting for laboratory procedures is impractical, especially while assigning putative functions to hypothetical protein data and lead discovery. Nonetheless, protein localization information unlocks other important biological information like a primary understanding of protein function, interacting partners, post-translational modifications, and signaling cascade, which are essential for a wide range of life science projects and during drug discovery. Henceforth, computational programs are built to acquire such data.
Amino acid composition, Motif, Domain, Interacting Partners, Homology information [through BLAST searches], Position-Specific Evlutotionary profiles [Scoring Matrices (PSSMs) & BLOSUM (BLOcks SUbstitution Matrix)], Gene Ontology (GO) terms and other associated data assembled in various databases and literature are utilized as datasets to develop protein location prediction tools. Support Vector Machines (SVM), the Bayes method, decision tree, k-nearest neighbor (KNN), Kernel-based logistic regression, Random Fields, and Neural Network/Deep Learning are the computational approaches implemented by these tools to calculate probable protein location. Among them, MULocDeep possesses advantageous features owing to its advanced framework.
MULocDeep
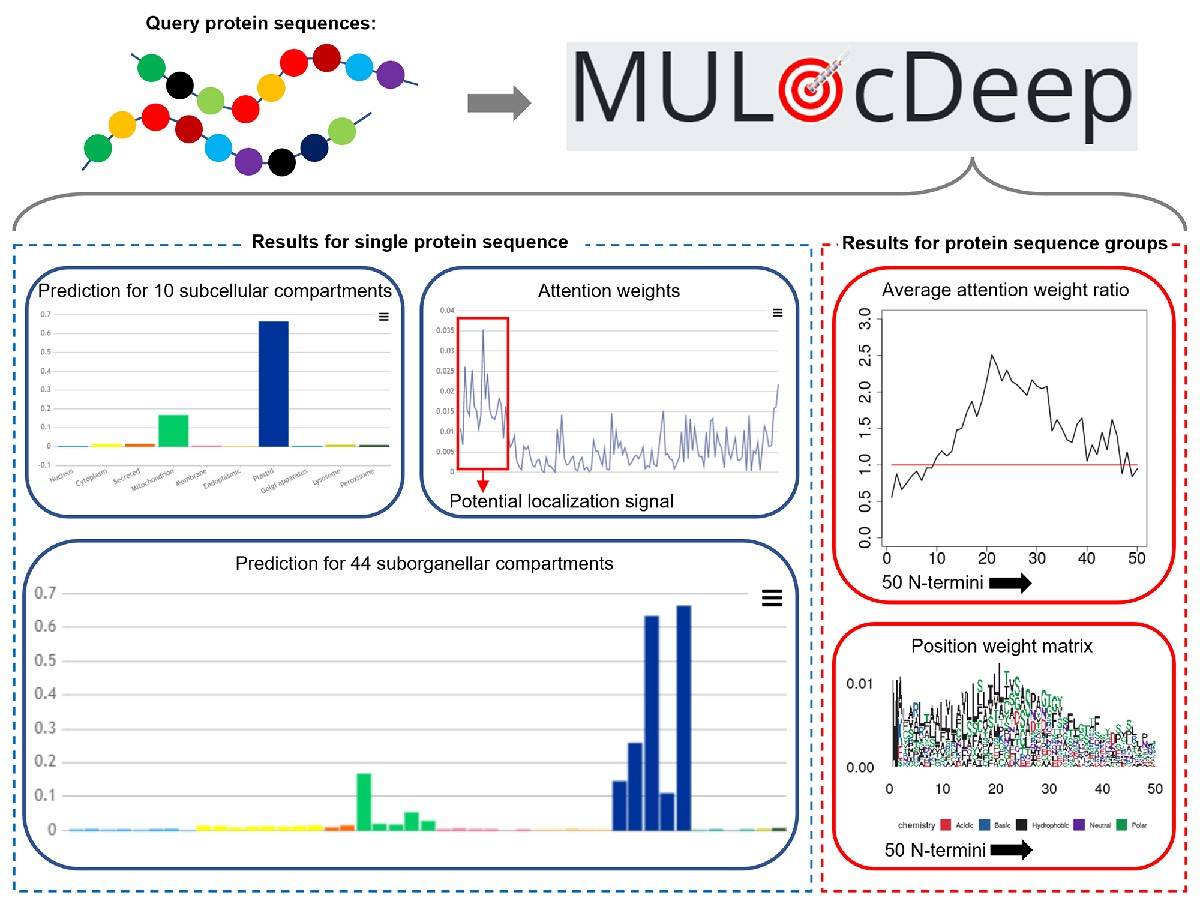
The developers of MULocDeep designed its previous version, MU-LOC, in 2018 for predicting plant mitochondrial target proteins based on N-terminal pre-sequences, internal sequences, and functional features, implementing a deep neural network and support vector machine. It outperformed six state-of-the-art tools for plant mitochondrial targeting prediction. MULocDeep web service provides a multi-label protein localization framework that implements bidirectional LSTM (Long Short-Term Memory) for handling protein sequences and a multi-head self-attention mechanism that assigns weights to each amino acid. It provides several benchmark datasets, including the Mito3 dataset and a comprehensive dataset for 44 suborganelle protein localization from the UniProt database (UniLoc dataset).
One protein may be localized into different cellular locations in its lifespan (multi-label). Most of the available prediction tool built using deep learning fails to address this aspect. MULocDeep addresses this shortcoming. Developers designed a matrix layer to apprehend the intrinsic hierarchical relationships between organelles and their subcompartments to offer predictions at both subcellular and suborganelle levels simultaneously and used two layers of bidirectional Long Short Term Memory (LSTM) and multi-head self-attention framework to extract biological signatures contributing to localizations at the resolution of single amino acid. Moreover, the model illustrates the contribution of each amino acid in localization and suggests localization motifs through the attentive embedding of assigning higher weights to specific parts of a protein sequence involved in protein regulation and thus detects the sorting signals from termini as well as internal sequences.
For the latest version, three species-specific MULocDeep models for fungi, metazoa (animal), and viridiplantae were trained using a transfer-learning approach to acknowledge the fact that targeting mechanisms often vary in different species. This variant outperforms the original version. Moreover, compared with DeepLoc2.0, the species-specific MULocDeep had a higher accuracy, higher or similar MCC (Matthew’s correlation coefficient) in ER, and higher MCC in the Golgi apparatus. The variant MULocDeep method is competitive with state-of-the-art methods.
MULocDeep’s interpretation of attention weight is not only attributed to signal peptides but also other biological significance of protein sorting, even though such labels were not applied to train the attention layer. Apart from species-specific features, the latest version offers parallel execution of multiple prediction tasks and population-based sorting mechanism analysis and visualization for query sequences, conforming to industrial standards. The web service is quite user-friendly, and the visualizations of sorting mechanism computations can be downloaded as publication-ready figures.
Conclusion
The upgraded protein localization prediction tool, MULocDeep, brings several new features and advancements to the field. The researchers firmly believe that mindful application of the model can be favorably utilized for medicinal research and Drug development studies. Along with human-related studies, the upgraded MULocDeep will certainly assist researchers in obtaining more accurate results while exploring other organisms as well. As proposed by the developers, we can expect more upgradation based on implementing Graph-based neural networks for feature representations and prediction, extending datasets to enable in-silico disease-specific mislocalization studies, and refinement of confidence assessment.
Article Source: Reference Paper | MULocDeep: Web Service
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.