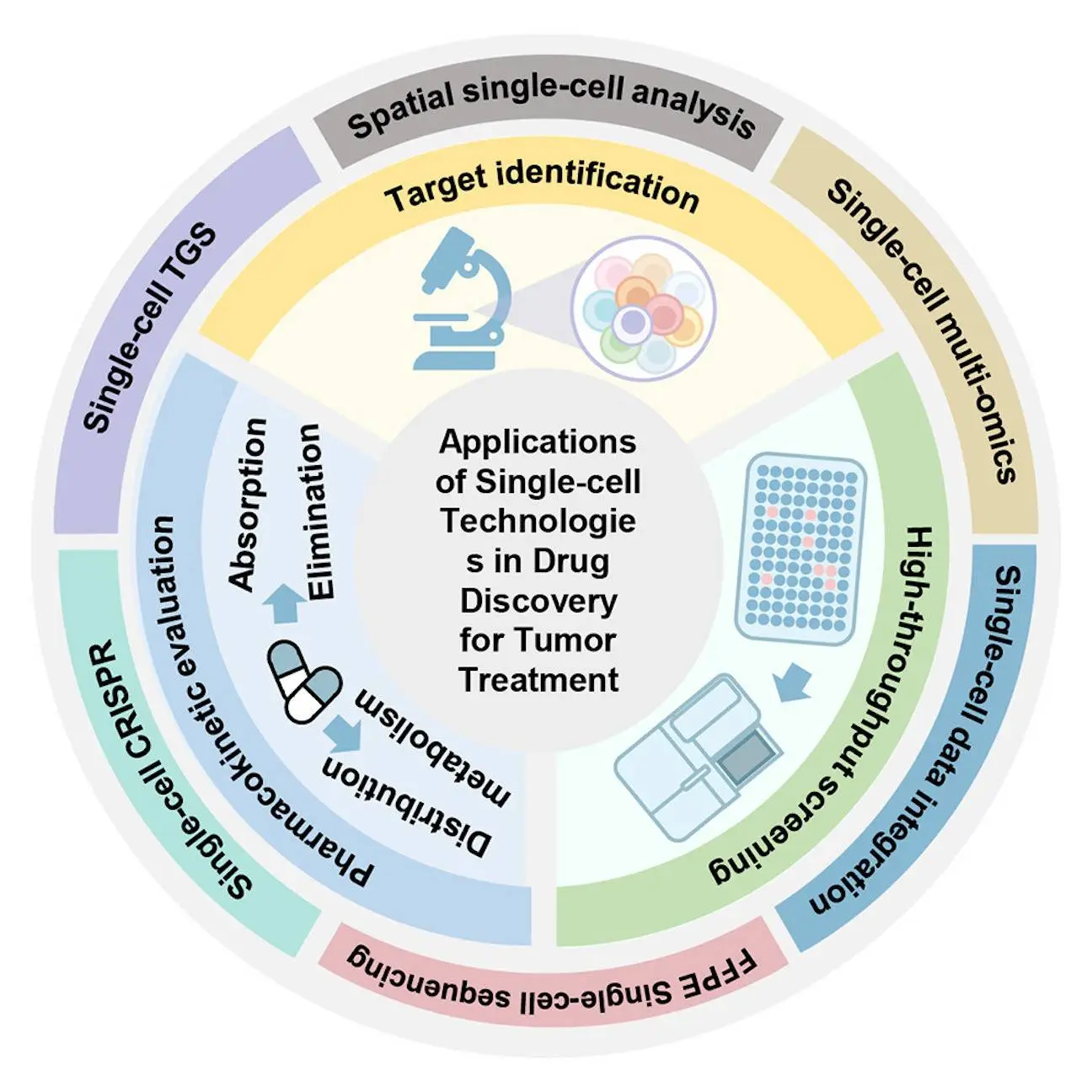
Single-cell technologies are effective drug development and discovery tools, serving important functions at several phases. They aid in identifying potential therapeutic targets, streamline single-cell high-throughput screening, and support pharmacokinetic investigations of anti-tumor medications. New tools for single-cell research combine multiple molecular information, increase the size of data, and characterize the influence of genes on phenotypic outcomes. Researchers outlined emerging single-cell technologies in this review, which offers a thorough understanding of tumor biology. They also methodically summarised the applications of single-cell technologies in various sections of drug discovery for tumor treatment, including target identification, high throughput drug screening, and pharmacokinetic evaluation. It is anticipated that drug discovery based on single-cell technologies will enhance therapeutic approaches and enhance the clinical results of cancer patients.
Introduction to Single-cell Technology
Single-cell technologies, including transcriptomic, proteomic, epigenomic, and genomic technologies, offer a comprehensive understanding of biological systems with single-cell resolution. Large cell populations are enhanced by these technologies, which also show the variety of tumor tissues and paint a thorough picture of intricate biology. Nevertheless, certain cell subpopulations and states that are crucial for tumor growth and treatment response may be hidden by the signal averaging process. Despite this, single-cell technologies are able to examine the cellular state, interactions, and regulatory mechanisms of tumor cells and their surrounding environment at a level down to the single cell, offering valuable insights into important aspects of the development of anti-tumor drugs, such as the evolution of tumors, the development of therapeutic resistance, and the mechanisms of metastasis.
Single-cell Technologies in the Treatment of Cancer
Single-cell technologies have completely changed how tumors such as lung, breast, and gliomas are diagnosed and treated. These technologies provide light on drug resistance mechanisms, genetic subtypes, and tumor heterogeneity. They analyze the nature of the tumor microenvironment and detect uncommon subpopulations such as tumor stem cells and circulating tumor cells. These methods enhance drug resistance in the treatment of tumors and aid in identifying possible therapeutic targets for drug candidates. By gaining knowledge about tumor biology, drug disruption, and pharmacokinetic effects in cells with varying genetic backgrounds, single-cell technologies also help researchers comprehend drug responses and ADME mechanisms at the cellular level.
Single-cell Technologies in Drug Development
Drug development greatly benefits from single-cell technologies, which make it possible to identify the mechanisms underlying drug resistance in tumor cells. By cutting expenses and shortening the development cycle, these technologies can increase the effectiveness of drug development. In pharmacokinetics research, they can also optimize lead molecules, which raises the success rate of medication development. Single-cell technologies improve drug development even more when patients in clinical trials are routinely monitored, identifying drug resistance mechanisms and screening biomarkers for response to therapy.
About Single-cell Third Generation Sequencing
Understanding the cellular state and functional regulation of tumors has been made possible by the extraordinary resolution with which ScRNA-seq has revealed gene expression levels in individual cells. Nevertheless, most current scRNA-seq coverage is limited to reading counts of polyadenylated transcripts 3′ or 5′ ends; mRNA splicing is not covered. By detecting full-length cDNA and RNA, TGS technologies—such as single-molecule real-time (SMRT) sequencing and nanopore sequencing—overcome this restriction and make it possible to identify complex DNA structural variants, whole transcript selective splicing events, and cell-type-specific mRNA isoform expression.
Extrachromosomal DNA and structural variations linked to tumor cells have been found using single-cell TGS technologies, such as RAGE-Seq and Nanoranger, in several investigations. While patients with acute myeloid leukemia employed Nanoranger to increase the resolution of leukemia and immune cell morphologies, RAGE-Seq was used to identify complete antigen-receptor sequences in breast cancer patients. Lower sequencing depth, however, may result in data sparsity and less accurate results by reducing the accuracy of single-cell TGS analysis. While sophisticated computational techniques such as scNanoGPS have been created to optimize the results of single-cell nanopore TGS, more work is required to overcome frequent shortcomings in single-cell TGS.
Limitations
Using single-cell technology in drug development faces several drawbacks, such as sample quality sensitivity, low cell throughput, and high cost. Furthermore, because of the dynamic nature of gene expression and cell transformation in tumor development, a unidimensional single-cell analysis is insufficient to determine precise cell subtypes or lineages.
Conclusion
Single-cell technologies have completely changed how tumors such as lung, breast, and gliomas are diagnosed and treated. These technologies provide light on drug resistance mechanisms, genetic subtypes, and tumor heterogeneity. They analyze the nature of the tumor microenvironment and detect uncommon subpopulations such as tumor stem cells and circulating tumor cells. These methods enhance drug resistance in the treatment of tumors and aid in identifying possible therapeutic targets for drug candidates. By gaining knowledge about tumor biology, drug disruption, and pharmacokinetic effects in cells with varying genetic backgrounds, single-cell technologies also help researchers comprehend drug responses and ADME mechanisms at the cellular level. Future studies should concentrate on combining geographical data and multi-omics data to fully comprehend tumor biology and create novel therapeutic approaches.
Article Source: Reference Paper
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.