A recent paper authored by researchers from the University of California, USA, and the University of Cambridge, UK, introduced an open-source Machine Learning framework, Autoprognosis 2.0, to encounter the technical and practical challenges that are obstructing the large-scale adoption of Machine Learning (ML) models in clinical settings and to empower non-ML expert medical professional, facilitating the development of diagnostic and prognostic models. AutoPrognonsis2.0 is shown to surpass existing risk score models as inferred from evaluating risk prediction of diabetes using data from UK Biobank and has been implemented as a web-based decision support tool that is extensively accessible to clinicians and patients. The study implies that AutoPrognosis 2.0 can be utilized by medical professionals to design predictive diagnostic pipelines and new clinical tools.
Artificial Intelligence in Health Sector
Artificial Intelligence (AI) has already impacted biomedical sciences, and certainly, more breakthroughs will unravel in this decade itself. Machine Learning Models can deliver a data-driven approach to inferring clinical decisions, bringing feasibility to maximize the accuracy of personalized diagnoses and thus prospective in revolutionizing disease care and management system by emerging as fundamental clinical tools.
However, apart from legal and ethical factors that should be primarily taken care of by the legislative authority, the technicality associated with the applicability of Machine Learning systems is one of the major reasons for the hindered prevalence of Machine Learning models in the public healthcare sector for disease diagnosis and prognosis. The researchers continuously approached to overcome the challenges.
The research group described the AutoPrognosis framework in a 2018 publication, which successfully applied it to design prognostic models for breast cancer, cardiovascular disease, and cystic fibrosis. The improved version AutoPrognosis 2.0 resolved the shortcomings in the algorithmic and usability perspectives of the previous framework.
Democratizing Machine Learning Models with AutoPrognosis 2.0
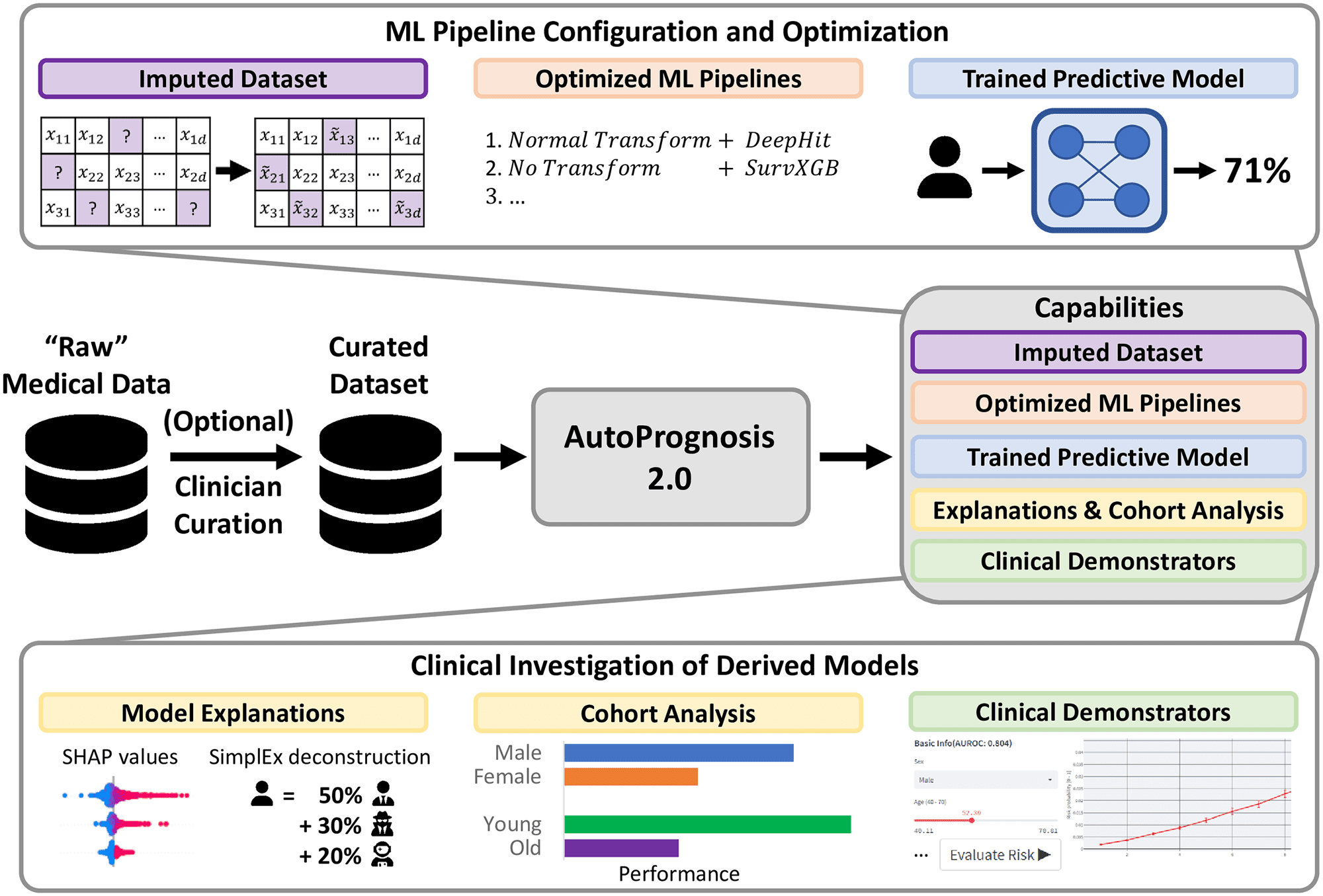
AutoPrognosis 2.0 aspires to make current statistical and machine learning methods accessible to ML experts and non-expert medical practitioners and thus overcome major issues accompanied by integrating ML models into diagnostic facilities. AutoPrognosis 2.0 configures and augments ML pipelines leveraging cutting-edge advancements in automated machine learning (AutoML) to devise robust predictive models, alongside it incorporates model interpretability tools to guide users, even who are with little technical expertise, in understanding operations as well as debugging the models.
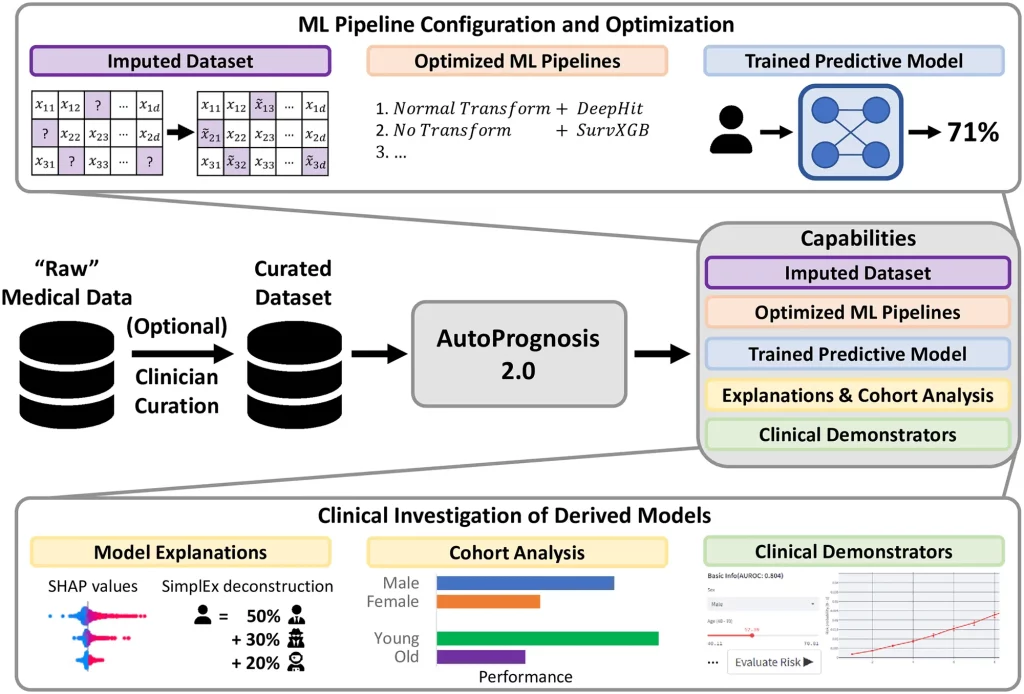
Image SImage Source: https://doi.org/10.1371/journal.pdig.0000276
The algorithm and software package enables clinicians to develop disease-specific diagnostic and prognostic models with only a minimal acquaintance with R or Python programming language. AutoPrognosis 2.0 can democratize ML as it automates ML pipeline optimization that involves data processing, model development, and model training and conceivably appears as a boon uplifting lives.
The framework manages entire steps in the computational operation, including missing data imputation, feature processing, model selection and fitting, model interpretability or explanations, and clinical demonstrator production when a user has specified a suitable cohort of patients and a desirable outcome. It is competent enough to ensure an appropriate and optimal selection of methods and hyperparameters relevant to each level, irrespective of inputting unnecessary, inappropriate information. Henceforth, encourage clinicians to fully utilize technological advances by downsizing the prerequisite technical proficiencies.
Overcoming Challenges in Deploying Diagnostic and Prognostic Models with AutoPrognosis 2.0
AutoPrognosis 2.0 implements an AutoML approach to automate the process of pipeline configuration by navigating a broad algorithmic search space efficiently and systematically conducting missing value imputation, feature processing, model selection, and hyperparameter optimization. Therefore, AutoPrognosis 2.0 fulfills the lack of a powerful ML Pipeline for clinical purposes as it can make multiple interdependent choices, in an unbiased manner, including imputation strategy, data preprocessing methodology, choice of the best model, configuring hyperparameters, which usually require technician intervention.
For instance, medical datasets are often incomplete; imputation is necessary to ensure the data is complete. Keeping this in mind, the framework is included with eight common imputation algorithms to allow users to select an appropriate imputation method. Additionally, the model is augmented with a state-of-the-art AutoML approach for imputation, HyperImpute, which automatically configures feature-wise imputation models. Further, it is capable of optimizing over five dimensionality reduction and six feature scaling algorithms.
As it consists of twenty-two classification algorithms, seven regression algorithms, and seven methods for survival analysis, it learns relationships between covariates to compute an optimized solution efficiently. AutoPrognosis combines the best-performing models into a single ensemble. The model possesses the ability to calculate transparent risk equations using symbolic regression from optimized models.
Furthermore, the model is eligible to overcome other important problems faced during the application of ML models. AutoPrognosis 2.0 allows interpretation of the advantages of ML-based methods over traditional approaches, such as Cox proportional hazard and linear regression models, and automatically identifies the best approach; at minimal technical cost. The model can determine the value of variable information critical to model development.
Seven state-of-the-art interpretability methods are incorporated into the model help users to understand and debug ML models. AutoPrognosis can help decide whether to update previous clinical predictive models as more data are assembled, and clinical practices are changed. Also, it addresses the challenge of transparency in reproducibility by providing a standardized, publicly available framework for training predictive models.
Conclusion
The researchers displayed the applicability of AutoPrognosis 2.0 in a practical scenario by executing prognostic risk score prediction for diabetes using a cohort from UK Biobank. It achieves greater discrimination for diabetes than expert clinical risk scores. Similarly, it can be applied to develop diagnostic and prognostic models for any clinical scenario and disease. The study defines a comprehensive overview of the challenges limiting the development and implementation of ML models in clinics and elaborates on the means to overcome them through the virtues of AutoPrognosis 2.0.
This novel endeavor of the researchers is simultaneously eligible to solve time-to-event problems; automate and optimize ML pipelines confining the most appropriate models and hyperparameters; provide feature-based, example-based, and closed-form risk equations; enable medical professionals to select variables and understand the value of information; and allow sharing the models across the clinical community. This open-sourced, integrated and automated framework can be utilized without much ML knowledge to develop models for more reproducible, optimized, personalized diagnosis, prognosis, and risk assessment.
AutoPrognosis 2.0 can potentially democratize Machine Learning in clinical practices, empower clinicians, medical researchers, biostatisticians, and epidemiologists and revolutionize the healthcare sector with more rapid, accurate, and efficient decision-making, disease identification, management, and treatment, benefitting crores of lives. Lastly, the users, i.e., medical practitioners, will be equally responsible for the effectiveness of this versatile framework because inappropriate data curation will lead to erroneous and devastating results. The researchers promise to upgrade the framework continually. Ultimately, regardless of the model, how rapidly these advancements are integrated with medical systems will decide the progression of the healthcare system.
Article Source: Reference Paper | AutoPrognosis 2.0: Software
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.