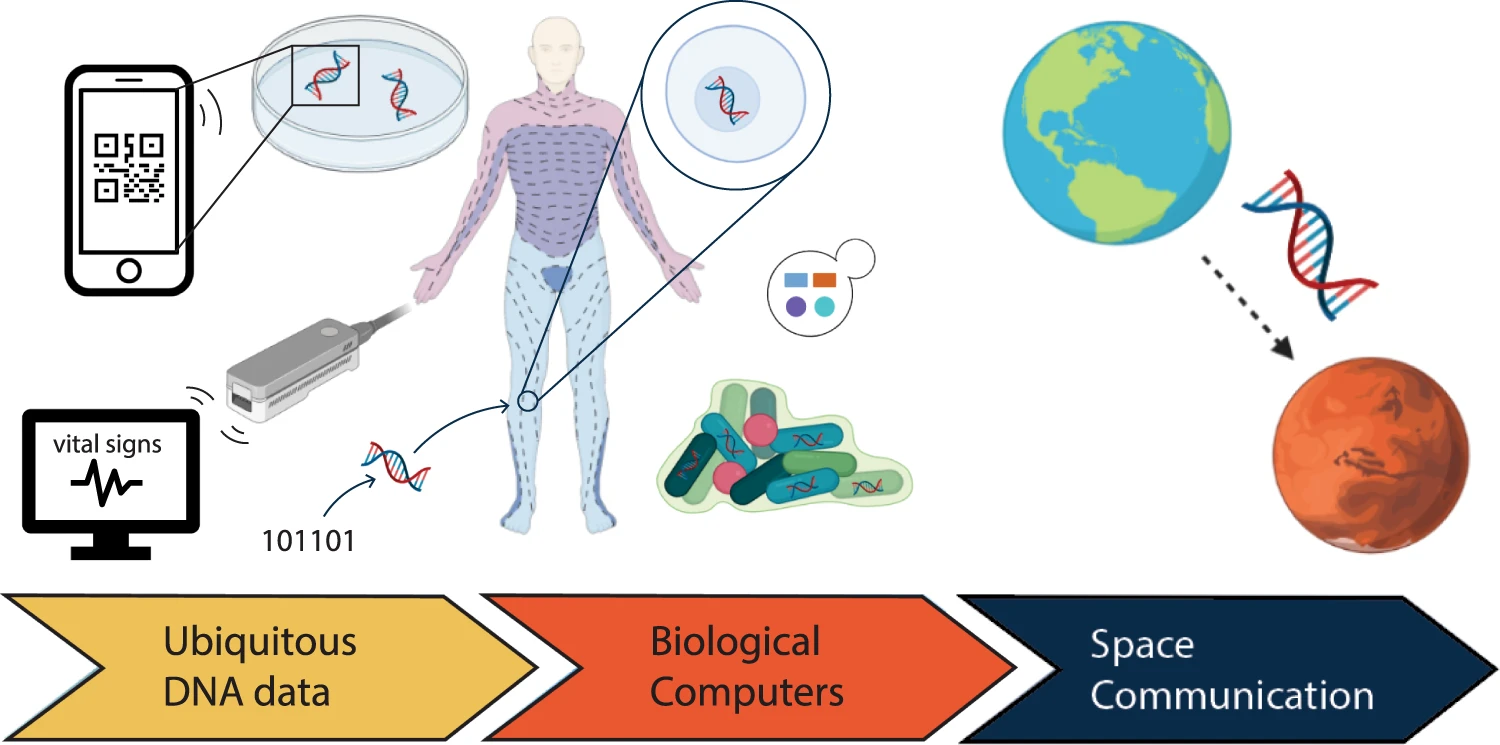
Synthetic DNA is becoming a popular alternative to electronic-based technology in areas including data storage, product tagging, and signal processing, etc.
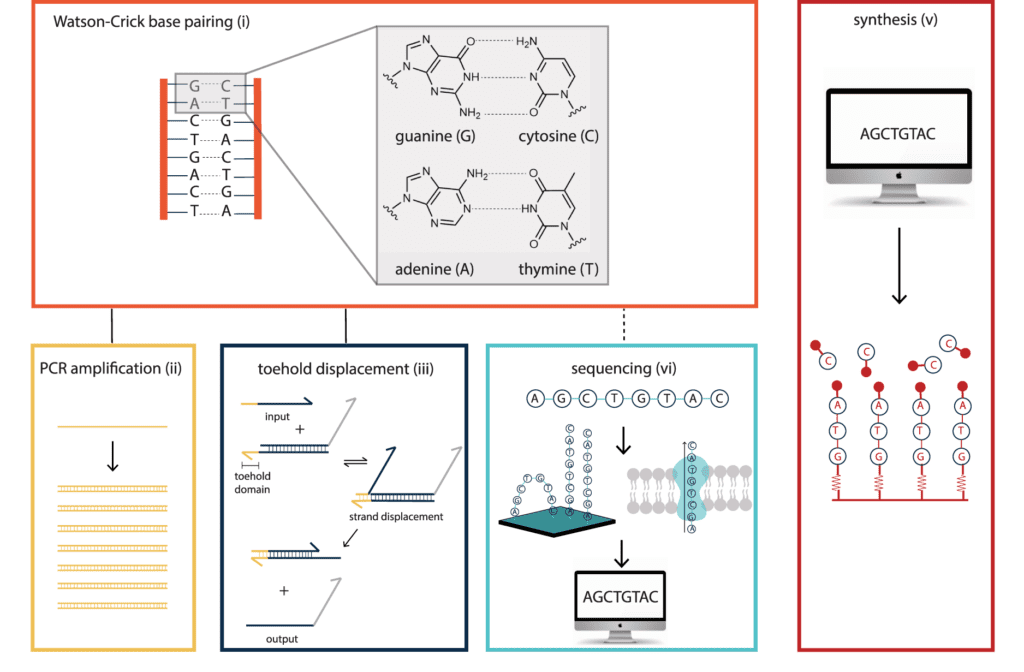
Image Source: Synthetic DNA applications In information technology
To date, DNA has shown to be the ideal material for use in information technology. Watson–Crick base pairing is a fundamental feature of nucleic acids that provides the foundation for various DNA handling methods, allowing for a broad range of computational applications across disciplines. These handling technologies were initially designed for the life sciences but have since been steadily repurposed. They are based on the biologically evolved properties of the DNA molecule.
The chemical business has been deeply influenced by improvements in information technology, mainly through the digitization and automation of chemical processes, with the pharmaceutical industry experiencing the fastest-growing demand. The chemical industry’s relationship with information technology does not have to be one-way, with information technology being used to help the chemical industry advance. Many possibilities open up when the chemistry is used to develop information technology, particularly to augment electronic systems with chemical systems.
Information Technology Applications of DNA
The biomolecule has found widespread application in information technology systems, thanks to innovative combinations of DNA handling methods. DNA has a high information density, can be chemically maintained for thousands of years, and is especially appealing for future advancements because it is the genetic material of all living organisms, making it eternally relevant for reading DNA.
Synthetic DNA has several applications that are thought to have the potential to affect the future of information technology.
- DNA as a Data Storage Medium
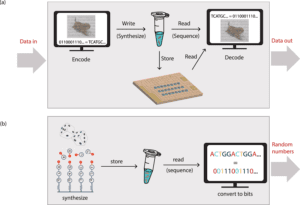
Image Source: Synthetic DNA applications in information technology
DNA offers a compelling alternative to traditional media for archival data storage. It has a data density that is more than six orders of magnitude higher than any mainstream storage medium, needs around eight orders of magnitude less energy at rest, and maybe kept for millennia when appropriately treated. The biodegradable nature of DNA molecules and DNA data storage being more environmentally sustainable than existing storage methods, these could be the key elements in DNA replacing magnetic tapes for archival data storage.
Using error-correction codes, a computer translates a string of bits to DNA sequences. This also adds redundancy, allowing the digital file to be retrieved and read later if the information is lost during the storing process, as well as an index to distinguish between the individual sequences. These DNA strands are next physically replicated in multiple copies using automated synthesis. The stored information can be protected from environmental variables by freezing the DNA strands in solution, enclosing the DNA molecules in microscopic silica particles, or by drying the DNA.
The DNA is amplified using PCR to recreate millions of copies of the DNA strands, which may then be read using sequencing methods to extract the digital content.
- DNA Barcoding and Tagging
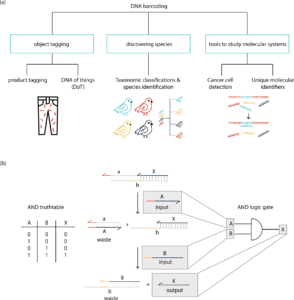
Image Source: Synthetic DNA applications in information technology
The barcoding with DNA is used to classify species, research molecular systems, and tag items, among other things. DNA product tags are DNA sequences that are introduced to a product for purposes of identification or traceability. DNA barcodes are very useful for tracing commodity products, and several commercial services that use DNA to confirm authenticity already exist.
Because DNA and many encapsulants are non-toxic, the detection limit can be as low as 0.001 ppm, making DNA ideal for product barcoding in these circumstances. If the value provided to a product by marking it is greater than the expense of incorporating DNA barcodes into the product, then the DNA barcode is particularly applicable.
The presence or absence of a synthetic strand, which allows binary classification of items, is the most basic DNA product tag. It is possible to detect the existence of such a product tag using PCR. By putting more information into the DNA sequence, DNA barcodes can grow exponentially richer in data.
DNA tagging in industrial applications to reduce counterfeit products has sparked a lot of attention. Several sectors have adopted these approaches for product control and supply chain responsibility therefore, DNA barcodes offer a far broader range of uses than merely product labeling.
- DNA of Things (DoT)
In 2020, the DNA of Things (DoT) was introduced, with uses ranging from data storage to object DNA barcoding. The idea is to 3D-print memory-enabled items by encoding the needed information into the DNA, enclosing the DNA in silica particles, and then merging DNA-bearing particles into a variety of materials that may subsequently be utilized for 3D printing. DoT combines DNA data storage technology with the concept of barcoding to add information to an object that isn’t visible to the naked eye. DoT may be synthesized by commercial producers and is comparable to DNA data storage.
- Random Number Generation
Automated DNA synthesis is a source for true random number generation (RNG). It allows for a 0.3 MB/s genuine random number output. DNA is an excellent source of entropy that is both air-gapped and orthogonal to other RNGs. This entropy can be used as a secure seed in a pseudo-RNG or directly. The technique for generating DNA random numbers begins with the mixing of the four nucleotides before they reach the chamber for solid-state DNA synthesis and generate DNA strands with no discernible nucleotide order. More than 7 million GB of randomness can be generated in a single synthesis run. The DNA strands can be amplified, sequenced, and converted to bits using PCR. In just a few hours, millions of distinct DNA strands may be created via automated synthesis. The vast number of DNA strands that can be synthesized cheaply in a short amount of time provides an excellent source of random numbers.
- DNA Cryptography
It came into play when researchers used both the sequence information in DNA strands and the high structural versatility of DNA. This enables secure communication technologies such as DNA origami cryptography (DOC), which generates scaffold DNA nanostructures for complicated molecular patterning. This scaffolding technique creates a massive design area, allowing for a critical size of 700 bits in theory. The DOC design also allows for differential access to specific parts of an encrypted message and, in general, ensures information confidentiality, integrity, and availability (CIA), paving the way for biomolecular next-generation information security.
- DNA Circuits and Dna Neural Networks for Information Processing
The system of information processing entails taking a signal in one form and changing it into another. DNA has the advantage that it can code for a wide range of signal inputs due to the specificity of Watson–Crick base pairing. On this foundation, work has been done to build biomolecular switches that do not require enzyme catalysis. The creation of DNA-based signaling cascades and logic gates has sparked considerable attention. Short oligonucleotides are used as input and output signals in DNA-based circuitry. Because the input and output signals of DNA logic gates have the same molecular morphology, multilayer circuits can be created using cascades of signals. Large DNA circuits with up to 130 DNA strands have been built using this pattern.
A DNA signaling neural network detects a pattern by comparing it to designs in its memory to determine which pattern it is most similar to. The network’s operations are broken down into individual chemical reactions, whose outcome is decided by the signal concentration of the respective input signals, with fluorescence serving as a readout.
- Automated Synthesis
Image Source: Synthetic DNA applications in information technology
It is essential for writing vast pools of DNA so that all the features can be physically handled that were earlier mapped from images into DNA. Because hybridization between query and target features is the key to successfully locating photos with similar content, Watson–Crick complementary base pairing, as well as PCR amplification, are essential for retrieving query images. The sequence of features in the image recovered is then read and decoded back to the digital file using high throughput sequencing.
In the future, cheaper and faster methods of synthesising and sequencing DNA will aid advancement in applications such as DNA data storage, DNA barcoding, DNA of things, and random number generation, among others. Today smartwatches record a limited set of people’s vital signs, but DNA can record all the bioprocesses of our bodies. Hence, the usage of DNA could someday bring a full paradigm change by molecularly monitoring everything that happens in our cells and organs. New means of creating, sending, calculating, storing, and reading digital information will eventually be enabled by further research in this sector.
Story Source: Reference Paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Swati Yadav is a consulting intern at CBIRT. She is currently pursuing her Master's from Delhi Technological University (DTU), Delhi. She is a technology enthusiast and has a keen interest in the latest research on bioinformatics and biotechnology. She is passionate about exploring new advancements in biosciences and their real-life application.