A toolshed for synthetic biology, metabolic engineering, and industrial biotechnology is available through the Galaxy-SynBioCAD portal. Using the tools and workflows available on the portal, one can create libraries of strains that produce the desired chemical targets, spanning a comprehensive metabolic pathway design and engineering process from selecting strains and targets, designing DNA parts to be assembled, to generating scripts to drive liquid handlers for plasmid assembly and strain transformation. All tools are compatible with SBML and SBOL, which are standard formats. A study at four different sites illustrates how pathways are designed and engineered by building a library of E. coli strains that produce lycopene. Benchmarking these workflows is also done on paths validated by experts and the literature. The top 10 pathways generated by the workflows retrieve the validated pathways 83% of the time.
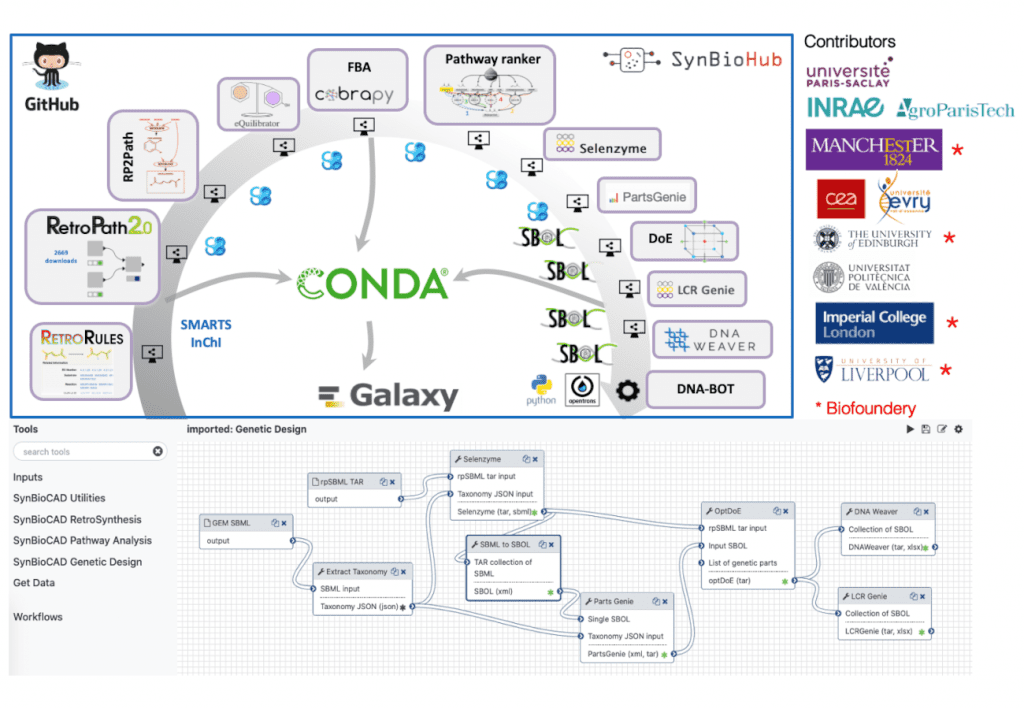
Image Source: https://www.jfaulon.com/galaxy-synbiocad-portal/
Life science research relies heavily on computation. Industrial biotechnology, metabolic engineering, and synthetic biology are not exceptions to this trend. A significant portion of this effort is devoted to developing workflows that adhere to engineering design principles such as standardization and abstraction of modular parts and decoupling design and fabrication. Appleton et al. review a number of design automation tools for genetic circuits that follow the electronic design automation concept (EDA). Cello is an example of a genetic circuit that uses EDA. Since Cello consists of several steps, input and output formats must be standardized. There are several formats for representing logic functions, including Verilog and Eugene, which encode sets of parts and constraints between them.
While Cello has been able to compile and standardize a number of genetic design tools, in general, this isn’t the case with the most freely available synthetic biology and metabolic engineering design tools, where fragmentation remains a major obstacle to their use. The past two decades, however, have seen the emergence of two main standards. Systems biology has developed SBML as a standard for encoding strains and pathways. Secondly, SBOL is a synthetic biology-specific data exchange standard. For the purpose of biodesign engineering, SBOL describes interactions between genetic components (DNA, RNA, and proteins).
A complex genetic circuit can now be encoded using SBOL, as well as metabolic pathways, vectors, and plasmids. An in silico genetic circuit design tool suite that conforms to SBML and SBOL standards has recently been proposed. The design of biosynthetic pathways can be assisted by a wide range of software tools. To summarize, the first step in determining the metabolic pathways between a target compound and its native metabolites occurs by finding metabolic reactions between the target compound and its native metabolites. For this step, retrosynthesis software tools are used, and if novel pathways should be identified or if compounds that produce unnatural targets should be found, reaction rules are used. The result of retrosynthesis is a metabolic map, and the pathways that link the chassis metabolites to the target must be enumerated in a second step.
Many pathways can be enumerated and searched using retrosynthesis software, which is sometimes integrated into it. Identifying the most promising enzyme sequences catalyzing metabolic reactions is the third step. Machine learning trained on metabolic databases can accomplish this through similarity searches to enzyme-annotated metabolic reactions. As a fourth step, pathways can be ranked after enzyme sequences have been annotated. There are many criteria for ranking that can be used, including thermodynamics, predicted yield of the target, flux balance analysis, cytotoxicity of the target and intermediates, and simpler criteria like pathway length.
Galaxy-SynBioCAD is a portal that provides computational tools for metabolic engineering and synthetic biology. Galaxy was selected as our workflow system because the tools in the ToolShed go beyond the original use of Galaxy for genome analysis. Proteomics, transcriptomics, metabolomics, flow cytometry analysis, and computational chemistry can all be cited based on the tool categories relevant to this study. As of 16 February 2022, Galaxy is used by several communities, producing many papers on omics (752 publications), microbiome (380 publications), cancer (386 publications), and drug design and discovery (96 publications). Galaxy has created a new category named ‘Synthetic Biology,’ containing 25 tools in the ToolShed.
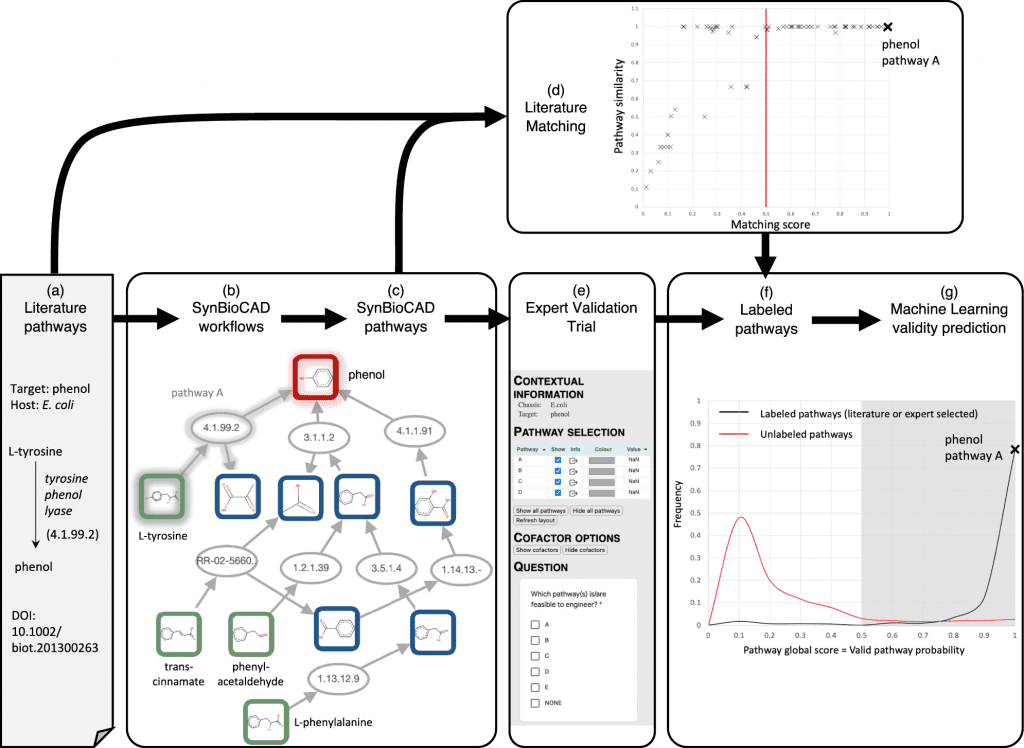
Image Source: https://doi.org/10.1038/s41467-022-32661-x
Among the features of Galaxy-SynBioCAD are tools for designing and engineering pathways. Galaxy-SynBioCAD is a community-driven initiative, which means our toolkit will continue to grow in the future. There are several software products listed in the introduction that could be included in the portal when it comes to pathway design tools. Using flux balance analysis tools, strains with knockout genes could be designed to maximize targeted product fluxes. A Galaxy workflow has already been created for processing and analyzing metabolomics flow cytometry data in the ToolShed, and these workflows can be incorporated directly into the portal to handle data generated during the ‘Test’ phase of the DesignBuild-Test-Learn (DBTL) cycle for synthetic biology.
There are also tools for genetic logic circuit engineering that could be considered. The global score of a pathway does not take into account the consumption of cell resources resulting from heterologous gene expression. Thus, it would be beneficial to integrate into the overall design pipeline a tool allowing for the prediction of metabolic burdens of genetic constructs based on enzyme expression levels.
Article Source: Reference Paper | Galaxy-SynBioCAD Portal
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.