Lives would not be made perfect without proteins since they are vital macromolecules that provide stability in their function. The study from Stanford University and Arc Institute presents a new concept known as ProteinDPO, a method that synchronizes generative models and performance landscapes with experiments in a bid to enhance stability predictions and protein design solutions.
The Promise of Generative Models
Generative models show potential for application in multiple areas, including the design and prediction of protein stability. These models, based on the principles of unsupervised learning, can encode various hierarchies and produce new sequences with the property displayed. The generative models are somewhat useful when it comes to designing proteins as they provide a way of searching through the space of sequences and predicting the stability, folding, and binding affinity of the protein. Their inherent capacity to work with big data and create diverse sequences makes them applicable in drug design and protein engineering processes.
Training Dataset
The datasets were used for the training of the protein generative models, validation, and optimization algorithms in the study. The Megascale dataset offered stability of protein variants, and the Homologue-free FireProt set offered ΔΔG of several mutations. The S669 dataset provided the actual costs from experimental alternatives ΔΔG for single mutations, and the dataset Native protein stabilities provided the free energy of unfolding for two-state proteins. Further, for the evaluation and training measures, we utilized the datasets of the SKEMPIv2 and AB-Bind. These various datasets allowed for a wide scope of experimental and theoretical examination of protein stability prediction and optimization algorithms in the work.
Generative Model Alignment
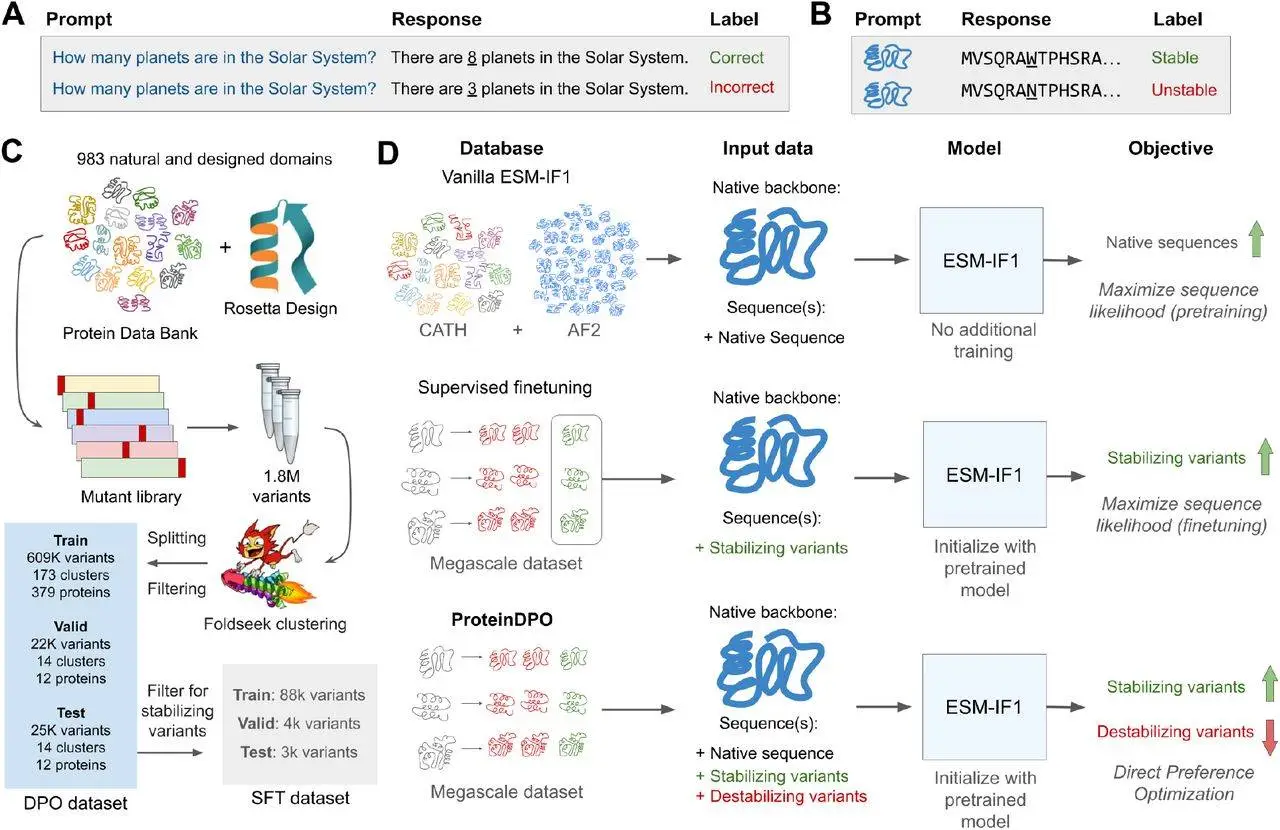
As for the generative AI applications, more typically in the natural language setting, training with feedback-based reinforcement learning (RL) in the post-training stage is common. Special attention should be paid to such an approach as it plays an important role in the successful outcome of these applications. The classical model training pipeline for generative models involves three main stages: supervised fine-tuning, feedback collection, and RL fine-tuning.
- Supervised Fine-tuning (SFT) where, in contrast to case-based retrieval, the model is fine-tuned on a positive example set selected due to its relevance to the property of interest. As applied to generative models of protein sequences, SFT employs samples of the highest quality, where the proteins feature superior stability or other desirable traits. When using SFT, the technique enables the model to optimize the likelihood over positively labeled samples, and in so doing, overfits the fine-tuning data and may lose generality from the information that was learned through the unsupervised pre-training phase. Thus, researchers have evaded the question of how this experimental fitness information could optimally be incorporated into protein generative models.
- Feedback stage: At the feedback stage, the model takes either the base model or the SFT model to create a plurality of potential completions for each input prompt from a set of preferences. Different ways of giving feedback include ranking comparisons whereby candidate outputs that are produced are compared based on the extent of preferences. The feedback stage is used to obtain evaluation scores from the generated candidate completions using some distribution parameterizations, which include the Plackett-Luce choice model so that the model can be motivated, improve its performance, and enhance its prediction based on the feedback given to it.
- Reinforcement learning stage: In the reinforcement learning stage, the second phase also undergoes feedback-based reinforcement learning to enhance its performance. Thus, instead of approaching a set of inputs and using a set of preferences to sample completions, the current model seeks to learn based on comparisons and rankings provided during the feedback process. Previous studies have employed comparison rankings in order to obtain evaluation scores and integrate those scores with the Plackett-Luce choice model to determine parameters that would help the model in its learning process. This stage is very important because it helps the final model bring in the feedback to enhance or tune the final output to meet certain goals and objectives.
Optimization Algorithms
For optimizing the objective, two primary algorithms are commonly used:
- Classical RL Algorithms include the Proximal Policy Optimization (PPO), which uses on-policy rollouts from the current model.
- Direct Alignment Algorithms: Such techniques include Direct Preference Optimization (DPO), which does not require on-policy data for training and only uses feedback data.
Direct Preference Optimization
DPO is a method that aims to adjust generative models according to properties of interest, which involves the provision of positive and negative examples. As for application in protein stability prediction, DPO is designed to enhance the stability corresponding prediction in the sense that the model is encouraged to select stabilizing variants rather than destabilizing ones according to measured experimental data. It is so exciting because it avoids overfitting the model and uses the full fitness landscape to improve the predicted level, for example, in cases where single mutations might dramatically change stability or conformation.
Weighted DPO Algorithm
The Weighted DPO is used to extend the Direct Preference Optimization (DPO) procedure, where each of the candidate sequences is assigned a numerical value to help the model identify the relationship between sequences. In another scheme, the option chosen improves the model by assigning weights to the scores and utilizing a regression-based model to redesign the model in line with the scores’ preferences. As seen, DPOw incorporates a means for efficient use of numerical data and increases the value of the model’s decision-making powers based on the scores given.
Benefits of Direct Preference Optimization
- Enhanced Model Alignment: Direct Preference Optimization (DPO) allows enhancing the generative models compared to the particular property to maintain the fit towards the generated traits by offering positive and negative preferences of similar instances.
- Preventing Overfitting: DPO also helps avoid overfitting to some correctly labeled samples through the simultaneous use of the entire fitness space, which enables the model to make robust and more generalized predictions.
- Improved Stability Predictions: For protein stability prediction, DPO, like ProteinDPO, can boost the stability prediction score by incorporating stability features into the pre-trained models, further producing robust and stable stability predictions across various protein domains.
Superior Stability Prediction: ProteinDPO Outperforms Existing Models
Direct Preference Optimization enables the incorporation of biophysical information regarding protein structuring into a structure-conditioned language model, which gives rise to ProteinDPO. This model can score effective stability and generate more stabilized protein sequences in comparison with the original and supervised fine-tuned models. Application to unseen protein structures: small protein monomers by training ProteinDPO on small protein monomers, they find that it generalizes to larger structures based on scoring the affinity and thermal melting points. As with other fine-tuning methods, such as supervised fine-tuning, ProteinDPO does not have such overfitting but instead remembers pre-training and native structure folding principles for proteins. Extensive evaluations with large datasets showed that ProteinDPO outperforms other models in stability prediction and can cope with many tasks more effectively. This approach can be applied to wider datasets and biological data than just protein.
Conclusion
In conclusion, ProteinDPO is a crucial advance for protein stability prediction and design, providing users with a versatile tool to help push the frontier of the protein language models and enable continued progress in computational biology.
Article Source: Reference Paper | Code for ProteinDPO is available at GitHub | Weights for the model trained with the paired DPO objective are available at Zendo
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.