Researchers from IIIT-Delhi and the University of Queensland have partnered to create Precily, a deep neural network-based modeling approach to predict the responsiveness of tumors to various drugs using gene expression data and anti-cancer drug descriptors.
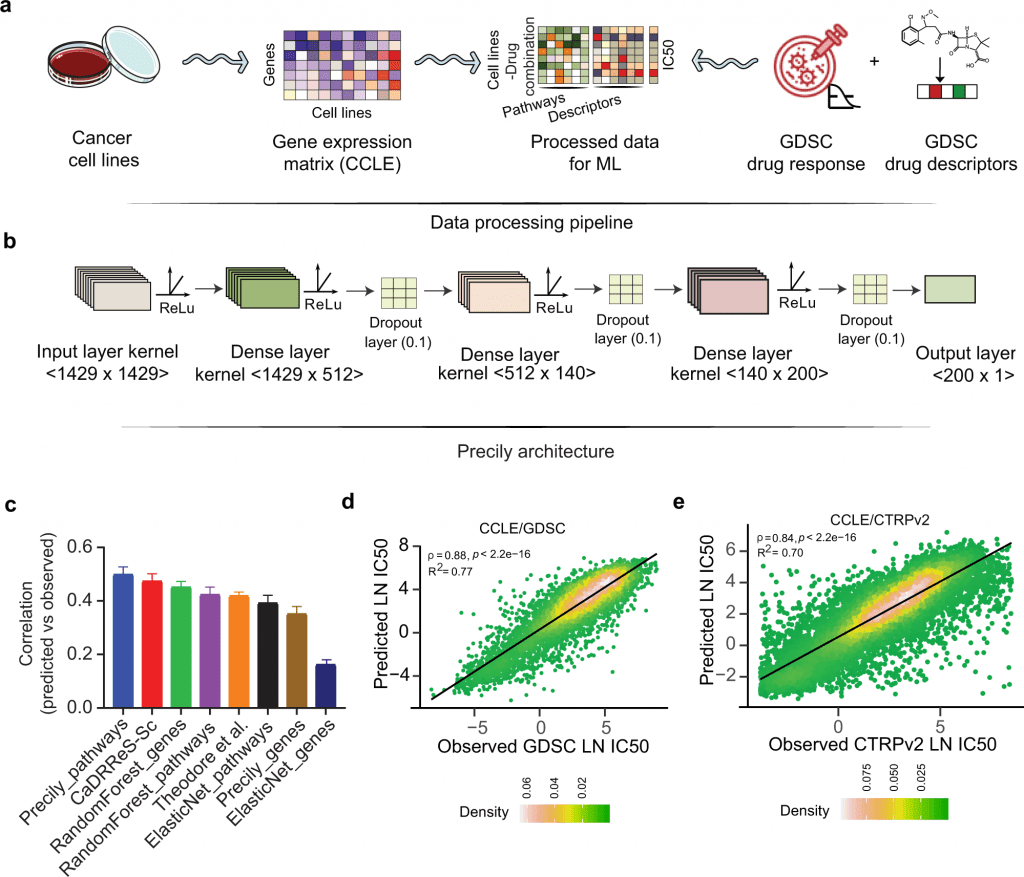
Image Source: https://doi.org/10.1038/s41467-022-33291-z
Personalized cancer medicine or precision medicine is the need of the hour for better treatment of highly convoluted diseases such as cancer. Tumor cells are known for their marked heterogeneity that stems from the molecular diversity of the cells present at the site of tumors. Treatment options such as Herceptin, which selectively targets HER2 (Human epidermal growth factor receptor-2) receptor-positive breast tumors, Afatinib for treating patients with EGFR (Epidermal growth factor receptor) gene mutations in lung cancer, and other various oncodrugs are available that suppress or inhibit differentially expressed genes and proteins.
Unfortunately, tumors can become resistant to the administered drugs, lessening the patient’s survival. Drug resistance occurs when a small group of cancer cells contains key molecular mutations, affecting the drug’s mechanism of action. Alternatively, the cancer cells can morph according to the drug’s action and make necessary molecular changes, thereby deeming their therapeutic effects insensitive. Thus, early predictions of a drug’s response in cancer patients exhibiting unique molecular diversity are essential.
Over the past few years, databases such as Cancer Cell Line Encyclopedia (CCLE), Genomics of Drug Sensitivity in Cancer (GDSC), and Cancer Therapeutics Response Portal v2 (CTRPv2) have been developed consisting of a wide array of genome sequencing data across thousands of cancer cell lines along with the information on anti-cancer drug sensitivity. The Cancer Genome Atlas (TCGA) is another landmark pan-cancer database consisting of clinical sample datasets with detailed information on patient’s genomic profiles and drug responsiveness. With the availability of such extensive knowledge, researchers have developed machine learning-based approaches to predict the response of a given tumor against the drugs. But these models have critical scientific gaps as they did not consider – 1) The structural properties of the drug and 2) The cellular pathways targeted by the drug. Therefore to address these issues, Precily was developed. Precily is a deep neural network (DNN) based framework that predicts the responsiveness of a drug by considering gene expression data, molecular descriptors of anti-cancer compounds, and pathway enrichment analysis that can provide insights into the cellular pathways and mechanisms of the query genes.
Findings
Precily was trained using the gene expression profiles from CCLE and the drug’s structural information obtained as SMILES notation. This enabled the DNN framework to suggest the sensitivity of the cancer cell lines against anti-cancer drugs with valid accuracy and reproducibility.
The researchers then tested the model to predict the drug responses in five prostate cancer cell lines. Precily highlighted the androgen receptor-positive LNCaP cell line to be most sensitive and the pathway involved in bringing about the sensitivity to be the PI3K/mTOR pathway which is directly related to cell proliferation and tumorigenesis; therefore, AKT and mTORC inhibitors that act by halting proliferation were predicted to have a marked sensitive response on the LNCaP cell line. Following drug selectivity, the model predicted drug sensitivity. It was found that prostate cancer cells showed a higher drug response to the therapeutics when they were in a highly active, proliferative state than in a quiescent state.
Precily extrapolated the drug sensitivity predictions for their applicability in xenografts. Xenograft helps us to model natural tumor environment and behavior. The LNCaP xenograft tumor in male mice was treated with Enzalutamide (ENZ), an androgen receptor antagonist widely used as a treatment option for castrate-resistant prostate cancer. ENZ acts by inhibiting cell proliferation and inducing apoptosis. With time, a cluster of samples exhibited ENZ resistance. Precily predicted that the genes associated with cellular proliferation seem to drive the resistance; hence hampering it by using an EGFR inhibitor along with the antagonist ENZ can provide better and more promising results.
Finally, the TCGA datasets containing the RNA-expression profiles from the patients were also investigated to ensure the model’s clinical relevance. The top drugs predicted by Precily can be used in tandem with other drugs, thereby bringing insights into combinatorial therapy approaches.
Final Thoughts
The study by Chawla et al. has successfully put forth a computational model named Precily, which proves to have a promising future as a primary screening tool in clinical trials. Precily has modeled and provided drug response predictions of cancer cell lines, xenografts, and patient data. It has also indicated that the drugs involved in treating obesity and type-2 diabetes can be repurposed as potential anti-cancer therapeutics against prostate cancer. Rich and valuable information on the combination therapy against drug-resistant tumors was also provided, although it needs further investigation. Given the limitations that Precily was unable to predict the efficacy of individual drugs and the lack of extensive cancer patient data with respect to drug responses, the results obtained were indeed encouraging as it takes us a step closer to greater accuracy in precision oncology.
Article Source: Reference Paper | Code availability: GitHub | Zenodo
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Shwetha is a consulting scientific content writing intern at CBIRT. She has completed her Master’s in biotechnology at the Indian Institute of Technology, Hyderabad, with nearly two years of research experience in cellular biology and cell signaling. She is passionate about science communication, cancer biology, and everything that strikes her curiosity!