The All of Us Research Program, with its ambitious objective of creating a diverse cohort of one million volunteers and making their data available for research, has enormous potential for revealing new information about human health and illness. Researchers have discovered more than 275 million previously unreported genetic variants, identified from data shared by nearly 250,000 participants of the National Institutes of Health’s All of Us Research Program. However, underlying this transformational movement is a complicated network of approaches for data gathering, processing, and access. This blog post explores the unique characteristics, potential impact, and bright future of genetic medicine of the program’s groundbreaking data release.
Introduction
“As a physician, I’ve seen the impact the lack of diversity in genomic research has had in deepening health disparities and limiting care for patients”
— said Josh Denny, M.D., M.S., chief executive officer of the All of Us Research Program
One of the main objectives of research on human health is to completely discover genetic diversity and record its role in health and illness, together with environmental and lifestyle influences. A major problem that has plagued the field of human genetics for decades is the under-representation of diverse perspectives in large-scale genomics studies. This biased viewpoint may have slowed the development of personalized medicine for all patients by restricting our understanding of how genetic variations impact health and sickness across groups. To close this gap, the All of Us Research Program, a groundbreaking initiative in the US, plans to make the largest and most diverse genome collection accessible to the general population.
Researchers used several data harmonization and quality control (QC) methodologies, as well as studies, to describe the dataset’s features, such as relatedness and genetic ancestry. They discuss the availability of WGS data from 245,388 ‘All of Us’ individuals, emphasizing the value of such high-quality data in genetic and health research. They also verified the findings by replicating previously reported genotype-phenotype relationships, including those for 117 other diseases and low-density lipoprotein cholesterol.
This information is made available through the All of Us Researcher Workbench, a cloud platform that combines and supports program principles, maintains ethical research practices, protects participant privacy through a passport data access mechanism, and provides equal access to data and computation.
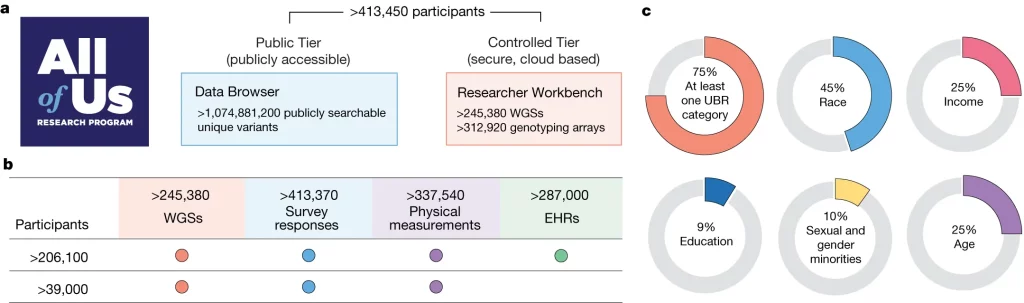
Image Source: https://doi.org/10.1038/s41586-023-06957-x
Mapping the Landscape of Human Variation
All of Us is dedicated to collecting and disseminating study data early and frequently to speed up health research. In 2018, the All of Us Research Program was launched with an ambitious objective of recruiting one million volunteers nationally, with a focus on historically underrepresented communities in biomedical research. This commitment to diversity is shown in the recently released data, which includes almost 245,000 individuals. Surprisingly, 46% of participants identify as members of racial or ethnic minorities, with 77% coming from underrepresented groups in research. Sequenced and genotyped individuals in this data release were not prioritized based on any clinical or phenotypic feature. Notably, 99% of participants with WGS data also have survey data and physical measurements, and 84% also have EHR data. This massive and diverse dataset provides a previously unprecedented opportunity to map the genetic landscape of human variety and represent the unique fabric of our common health narrative.
Building the Foundation: Participant Recruitment and Data Collection
The program started with a diverse cohort that was carefully assembled by reaching out to historically marginalized populations. Online informed consent encourages transparency and participant liberty. The gathering of data then coordinates a multimodal strategy:
- Electronic Health Records (EHRs): Using the OMOP Common Data Model, these medical history archives standardize vast clinical data, making cross-disciplinary analysis simple. Academics will always have access to up-to-date data, thanks to quarterly updates.
- Participant Surveys: Using the OMOP Common Data Model, these medical history archives standardize vast clinical data, making cross-disciplinary analysis simple. Academics will always have access to up-to-date data, thanks to quarterly updates.
- Biospecimens: Blood samples are mostly utilized to get DNA for genomic analysis, which enables scientists to look into the hereditary causes of both health and disease.
Opening the Gates: Data Access and Security
The All of Us program encourages responsible data access and open science. Applicants from a range of backgrounds can apply for a “data passport,” which acts as a doorway to participant data. This system finds a balance between rigorous security and free access:
- Six-step Authorization: Tight protocols ensure that researchers are qualified, that their projects are ethically sound, and that they adhere to data-use laws.
- Data Passport and Code of Conduct: To preserve participant privacy, a commitment to responsible data usage is required.
- Data Dissemination Policy: By restricting the dissemination of personally identifiable information, participant anonymity is promoted.
The Power of the Genome: Deciphering DNA
The All of Us program relies heavily on genomic data to unlock the secrets buried in our DNA. Whole-genome sequencing (WGS) offers a thorough comprehension of the genetic makeup of every individual. Careful methods, however, guarantee the accuracy and quality of the data.
- Stringent Sample Preparation and QC: DNA samples are meticulously gathered, processed, and inspected to make sure they meet quality standards before sequencing.
- Library Construction and Sequencing: DNA libraries are constructed and sequenced using specialized techniques, producing enormous amounts of unprocessed data.
- DRAGEN Pipeline for Analysis: This advanced tool ensures consistent data representation by precisely mapping, aligning, and calling variants.
- Standardized Processing: Regardless of the location of the sequencing, comparable data is guaranteed by maintaining consistency throughout all of our Genome Centers.
Beyond WGS: Complementary Tools for Specific Insights
WGS offers a broader perspective, whereas array genotyping has more specific applications. By focusing on pre-selected variants related to specific qualities or disorders, it provides:
- Faster and More Cost-Effective Analysis
- Quality Control and Confirmation.
Ensuring Data Integrity: The Art of Curation
Data is carefully vetted before it is made available to academics.
- Joint Call Set Creation: Merging WGS data from each member improves accuracy and identifies errors.
- Sensitivity and Precision Evaluation: Control samples offer dependable data for analysis, guaranteeing precise variant calling.
- Single-Sample and Joint Call Set QC: Multiple layers of testing look for contamination and irregularities in both the individual samples and the whole dataset.
- Batch Effect Analysis: Multiple sequencing sites yield consistent data quality thanks to extensive testing.
Navigating Relationships: Genetic ancestry and relatedness
The program detects individuals in the cohort who may be related. Genetic ancestry inference confirmed that 51.1% of the All of Us WGS dataset is derived from individuals of non-European ancestry. Briefly, the ancestry categories are based on the same labels used in gnomAD kinship scores are used to identify close relatives, and a “maximally unrelated set” is created for study. This guarantees:
- Accurate Research Findings: The absence of genetic similarities in the results enables the drawing of broader generalizations.
- Respect for Participant Privacy: Members of the immediate family take extra care.
From Genes to Traits: Unveiling the Connections
Polygenic Risk Maps (PGRMs):
These state-of-the-art tools combine the impacts of many genetic variations linked to a particular feature or illness. Researchers can ascertain an individual’s PGRM score by:
- Estimate a person’s genetic risk for various conditions: Assessing an individual’s genetic susceptibility to a range of ailments paves the way for individualized medical strategies that modify therapies and prophylactic measures by patient risk profiles.
- Explore how genes interact with the environment: When PGRMs are combined with environmental data, it becomes clear how different lifestyle choices and outside factors, based on genetic predisposition, affect health outcomes.
- Identify novel drug targets: By identifying the pathways regulated by genes associated with a particular disease, PGRMs can guide the development of customized drugs.
Genotype-by-Phenotype Replication:
The All of Us effort seeks to validate its discoveries while also making new ones. Researchers use PGRMs and other tools to replicate proven connections between genes and traits across several populations. This ensures that the results are not confined to certain groups and may be used generally.
From Promise to Reality- A Brighter Future for Genomic Medicine
Though groundbreaking, the All of Us initiative is far from finished. Researchers’ approach to doing human health research has undergone a paradigm change thanks to the All of Us Research Program. It clears the path for a day when everyone will genuinely benefit from genetic medicine by placing a high priority on diversity and free access. This varied dataset has the capacity to:
“The All of Us dataset has already led researchers to findings that expand what we know about health – many that may not have been possible without our participants’ contributions of DNA and other health information. Their participation is setting a course for a future where scientific discovery is more inclusive, with broader benefits for all”
— said Josh Denny, M.D., M.S., chief executive officer of the All of Us Research Program and an author of the study.
- Find novel pharmacological targets and therapeutic approaches: Through the identification of distinct genetic variants linked to illness, scientists may create more specialized treatments for a wider range of patients.
- Reduce health disparities: It may help create treatments that specifically target the needs of underrepresented populations by comprehending the genetic foundations of health inequalities.
- Encourage people to take responsibility for their health: Giving people access to their genetic information and educating them about how it relates to their health concerns
- Addressing Ethical Challenges: The program’s ethical sustainability depends on ongoing discussions about informed consent, data privacy, and equitable access to research benefits.
Conclusion: A Legacy of Open Science and Discovery
All of Us’s clinical-grade sequencing not only makes research possible but also provides participants with value in the form of clinically relevant genetic data and health-related features for those who choose to receive them. It is anticipated that in the next years, this collaboration with All of Us participants will allow researchers to go beyond large-scale genome discoveries and begin to comprehend the implications of applying genomic medicine on a wide scale.
Article source: Reference Paper | Reference Article
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.