Uni-RNA, a context-aware deep learning model developed by DP Technology, Beijing, China researchers, delivers state-of-the-art performance in unwrapping structural, functional, and evolutionary information intercalated in RNA sequences. The study has illustrated the dramatic potential of Uni-RNA, empowered by large pre-trained models, in fostering further understanding of the RNA world, paving breakthrough findings and developments.
A Glance into the RNA Network
The contribution of RNA molecules in sustaining cellular processes has brought significant attention to research regarding RNA. Supporting and mediating the central dogma of life, decoding DNA sequences meaningfully into proteins, is facilitated by coding RNA machinery, namely mRNA, rRNA, and tRNA. Apart from this conventional view of RNA functions, the discovery of different non-coding RNAs that don’t fetch any protein translation has added an unexplored perspective and dimension in cellular dynamics, evidencing the role of RNAs in regulating activities by orchestrating a plethora of functions.
The microRNA (miRNA) are non-coding small regulatory RNA that can modulate post-transcriptional gene expression and exerts an essential regulation role in different physiological and pathological processes in organisms. Small nuclear RNA (snRNA) facilitates the formation of spliceosome complexes mandatory for the removal of introns from pre-mRNA. Another group is long non-coding RNA, or lncRNA plays a role in differential processes by regulating allelic expression, guiding chromatin modification, transcriptional regulation, and RNA processing.
Study of RNA: The Associated Challenges
From gene regulation to catalytic activity, the outstanding diversities of roles committed by RNAs bring inquisitiveness to dissecting the underlying mechanisms and interactions for unraveling important phenomena and a comprehensive understanding of the cellular landscape. Comprehending the secondary and tertiary structures of RNAs, especially non-coding RNAs, is difficult owing to their dynamism, flexibility, and adaptability of conformations under different cellular landscape essential for executing a diverse range of functions. Also, understanding their intricated functions is also linked to their 3D structures and transient conformational transition.
This dynamism is difficult to capture through analytical techniques. In fact, the basic concept of comparative biology can’t accommodate and determine the structural and functional properties of RNA by investigating homologous sequences like done for proteins. In simpler terms, RNAs with unrelated sequences can exhibit similar functionalities. Due to the progression of sequencing technology, obtaining RNA sequences is not a big deal for researchers, but unlocking relevant information from that data is challenging due to the intrinsic high-dimensional complexities. The researchers have highlighted the need for unified and adaptable Computer-aided techniques to counteract the problems.
Uni-RNA and its Advantageous Features
The researchers have developed Uni-RNA anticipating to circumvent the above constraints regularly faced by scientists. The context-aware deep learning models are built leveraging BERT architecture, integrated with advanced techniques including rotary embedding, flash attention, and fused layernorm for optimal performance, and performed pre-training using one billion RNA sequences from different species and categories. The pre-trained large amount of data is important for learning and generalizing features and representations that can be fine-tuned or adapted for specific tasks more efficiently.
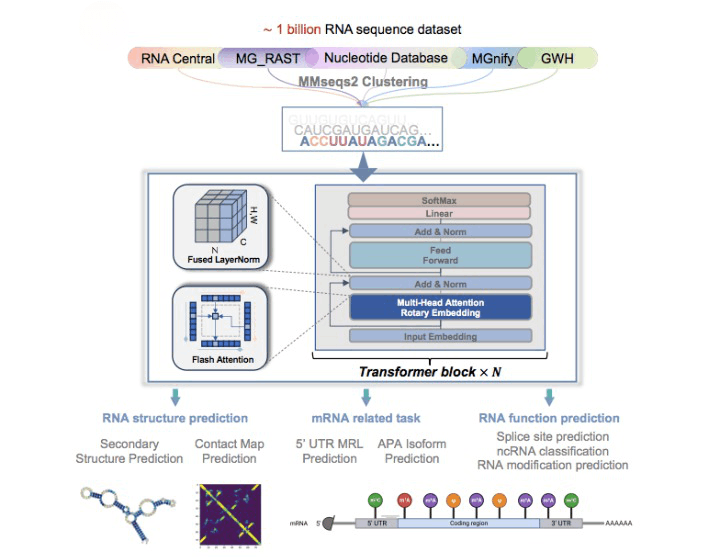
Image Source: https://www.biorxiv.org/content/10.1101/2023.07.11.548588v1
Although pre-trained models have received criticism, the authors believed that by employing appropriate mathematical descriptions for extensive data and sophisticated model architectures, the pre-train approach could capture the complete landscape of RNA. After fine-tuning Uni-RNA on various downstream tasks related to RNA structure and functions, it can achieve cutting-edge results. The evaluation studies reflect the following superior characteristics of Uni-RNA.
- The Uni-RNA demonstrated outstanding performance compared to the 13 models and previous SOTA (State-of-the-art) model in predicting RNA structures, a key to obtaining insights into RNA folding mechanisms, dynamics of RNA-protein, RNA-ligand interactions, and the development of RNA-based therapeutics. Uni-RNA extracts hidden information from billions of RNA sequences enabling highly accurate RNA secondary structure prediction.
- In comparison to the commonly used one-hot encoding for predicting RNA contact maps, the predicted structures obtained by the Uni-RNA model better captures long-range dependency relationships. The superior performance is perhaps attributed to the latent structural and functional information extracted by the pre-trained model.
- Moreover, it displayed accurate and robust performance in 5’UTR (UnTranlated Region)-based Mean Ribosome Load (MRL) prediction essential for analyzing translational activities and protein synthesis levels of mRNA under specific conditions. This can be applied to developing mRNA-based therapeutics.
- With respect to the previous SOTA, the Uni-RNA also achieves superior performance in Alternative polyadenylation (APA) isoform prediction based on 3’UTR Sequences. APA phenomenon is linked with generating mRNA isoforms and modulating gene expression levels.
- The model can also predict splice sites on pre-mRNA sequences leveraging the pre-training augmented significant generalizability capabilities. Understanding splice sites holds importance in various studies related to genomics and proteomics.
- Uni-RNA offers a computationally efficient sequence-based approach to classifying small non-coding RNAs that holds the possibility of inferring evolutionary information.
- Uni-RNA demonstrates incredible predictive performance across 12 artificial and natural RNA modification categories, exhibiting significant performance improvement compared to the commonly used one-hot encoding method. Understanding Post-transcriptional RNA modifications can assist in elucidating the functional implications and regulatory mechanisms underlying RNA molecules.
Conclusion
Multiple aspects of the extensively wonderful range of tasks meticulously performed by different RNAs are yet to be understood. Several computational approaches have been attempted and implemented before to address these aspects. In this regard, Uni-RNA offers a comprehensive solution to different sets of inquiries that are encountered by the scientific community. The efficient extraction of embedded features in RNA sequence by Uni-RNA will certainly empower research fields. From predicting tertiary and secondary structures, deciphering associated functions, and distinguishing different RNA classes to assisting in studies of developing mRNA-based therapeutics, Uni-RNA achieves supreme performance in all aspects. Henceforth, it could provide a universal solution for venturing deeper into the RNA universe. The researchers anticipate that Uni-RNA will help in decrypting the enigma of life, endow researchers with the ability to overcome challenges and enable breakthroughs.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.









