Scientists at King’s College London, UK, and collaborators have introduced ‘Flow,’ an open-access web platform to perform bioinformatics analysis. Flow provides the capability to execute bioinformatics workflows through an intuitive web interface. It enables efficient storage and organization of the resultant files while facilitating streamlined data sharing.
Flow was built to overcome the difficulties historically faced in analyzing, storing, curating, and sharing various data and information related to bioinformatics and make it more accessible to biologists with a lack of computational knowledge on a large scale. It offers solutions to simplify data administration and encourages teamwork and collaboration between scientists to enable greater progress at a faster pace in this emerging interdisciplinary field of research.
Existing Problems with Data Organization in Biological Research
Biological data has accumulated at an exponential pace over the years, and most of the information has been left scattered and unorganized, making it increasingly difficult for scientists to properly analyze key pieces of information when performing their research. Additionally, it is difficult for scientists to find the time to manage their own data, and they lack the expertise for it as well.
Some of the concerns brought up have been related to inadequate information related to data processing, the quality of data, and the accessibility of various platforms to obtain the required data. Currently, biological data and information related to bioinformatics are stored in databases such as GEO or ArrayExpress. The issue here is that only raw data that has been completely processed is stored, isolated from the analysis, and parameters that give information on the processes carried out to give the obtained result. With an increasing amount of data piling up day by day and considering the variation in experimental methodologies applied to produce the data, it is difficult to keep up with manually curated databases.
A ‘reproducibility crisis’ or ‘reproducibility iceberg’ has been historically observed, wherein there are very few manuscripts and pieces of literature that give both data and code for an experiment. In the few cases where both are provided, difficulties are faced in installing software and compatibility issues with the operating systems of the user’s device. Some information regarding metadata and parameters may be left incomplete. Such issues with reproducibility and portability have held up significant barriers for scientists from performing seamless, obstacle-free research.
What is Flow?
Flow is an open-access web platform created to overcome the aforementioned issues in an efficient and user-friendly manner. It creates a link between database solutions and information on analysis carried out using bioinformatics through a user-friendly and intuitive interface and web API. It can account for a wide range of methods used in genomics, and further, the addition of NextFlow pipelines used for bioinformatics analysis has been incorporated into the platform as well. Pipelines are used to integrate various programming languages and data related to protein sequences and execute the given information into a single, comprehensible pipeline; these pipelines are portable and can be reproduced for further studies as well.
This platform is specific to data exclusive to bioinformatics research and has made workflow much more convenient, easier, and cheaper than before. It is capable of running parallel tasks given to it and makes sure that the computer architecture remains the same regardless of where an individual chooses to run the code by executing code in ‘containers.’
Flow promotes the reproducibility of data as it is easy to access and use, and this is a significant contribution to research and development as, without it, it is impossible to move forward in any scientific study. It also raises the standards and quality of data accumulated on the platform, thereby increasing the quality of research carried out using it as well.
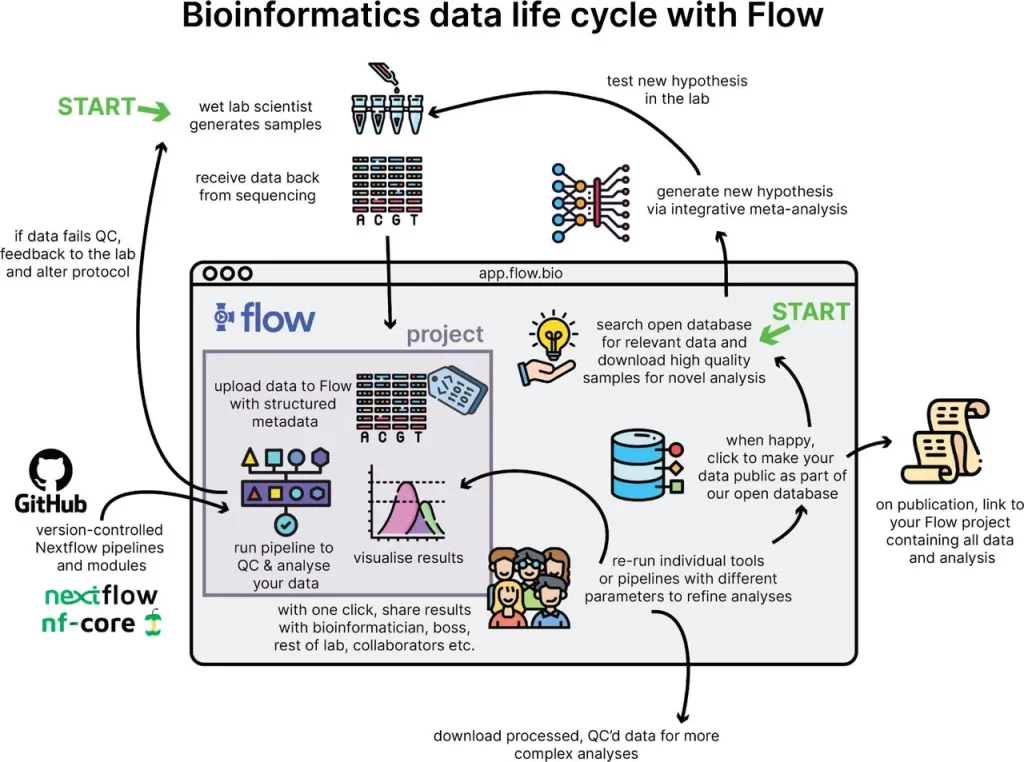
Image Description: Bioinformatics data life cycle with Flow. Users can enter Flow to analyze their own samples or search for public samples relevant to their research question.
Image Source: https://www.biorxiv.org/content/10.1101/2023.08.22.544179v1
Integration of NextFlow in Flow
The pipelines provided by NextFlow can be understood without much difficulty and enable community involvement, as anyone can contribute to the pipelines through GitHub and Slack. Since the utilization of NextFlow pipelines requires a prerequisite, basic-level knowledge in Unix programming, using them has been made easier by the introduction of Graphical User Interfaces (GUIs), such as Seven Bridges and form.bio. A disadvantage, however, is that these GUIs may be expensive for some users.
Flow has removed the drawbacks associated with NextFlow by combining it with solutions for web database management, along with a user-friendly GUI, at a much cheaper cost. Code and analysis parameters are provided right next to data objects, making navigation much easier and providing quality control. It is a democratic solution to sharing sequencing data, as the users don’t have to be part of a specific scientific organization to access or share information.
An overview of the architecture of Flow
There are five layers in the workflow, in a hierarchical order of organization:
- Bioinformatics layer: Here, the data obtained from bioinformatics data is processed and analyzed using a variety of command-line tools. The layers proceeding with this one provide additional inputs and increase the sensitivity of the information.
- Nextflow layer: The pipelines of Nextflow are organized here in a reproducible manner in the form of containers. It requires the installation of Nextflow and accommodates tools from the first layer as well.
- nextflow.py layer: The pipelines from the second layer are run on Python code while utilizing an extensive Python library. The resultant files are resolved into objects in Python for further analysis.
- API layer: A Django-based web app, flow-api, uses the third layer to interact directly with the pipelines. A PostgreSQL database is used to access data, which unravels a GraphQL API.
- Web client layer: Facilitation of interaction with Flow programmatically via the React Frontend takes place here. An official Python library called Flowbio facilitates this as well. It is anticipated that a majority of Flow users will end up using this layer, as it enables them to interact with the UI on the platform directly and does not require programming knowledge.
Features of Flow
Information related to RNA-seq, CLIP-seq, and ChIP-seq can be uploaded on the platform easily, as it supports a variety of file formats. Flow also enables users to track the history of data, detect any alterations that might have taken place, and retrieve earlier versions by implementing strong data versioning and origin tracking. Users can add information and annotations to their datasets by using the data annotation and curation tools included in Flow. Even with enormous volumes of data at its disposal, Flow is built to provide optimal performance, even when dealing with large-scale bioinformatics datasets. Within the same environment, researchers can analyze their data with more ease and comfort, owing to the platform being extensively integrated with bioinformatics analysis tools that are widely used.
Conclusion
The study introduced a web-based platform, ‘Flow,’ that integrates bioinformatic analysis and database solutions, addressing challenges such as data processing, quality concerns, accessibility, and curation in large-scale meta-analyses. Flow offers a user-friendly interface and web API, accommodating various genomics methods and allowing the addition of Nextflow pipelines via a JSON schema file. Bioinformatics research can reach greater heights than ever thought possible as long as relevant information is accessible to scientists, researchers, and developers. Platforms like Flow enable this by bringing greater accessibility by integrating databases with a plethora of sequencing information, therefore acting as a frontier for furthering research in bioinformatics.
Flow can be deployed on local systems and cloud services and can be freely accessed at https://flow.bio. It is currently managed at an OpenStack facility at King’s College London.
Article Source: Reference Paper
Learn More:
Swasti is a scientific writing intern at CBIRT with a passion for research and development. She is pursuing BTech in Biotechnology from Vellore Institute of Technology, Vellore. Her interests deeply lie in exploring the rapidly growing and integrated sectors of bioinformatics, cancer informatics, and computational biology, with a special emphasis on cancer biology and immunological studies. She aims to introduce and invest the readers of her articles to the exciting developments bioinformatics has to offer in biological research today.










