Scientists from Michigan State University have introduced the LongBondEliminator, a tool designed to detect and correct ring penetrations in molecular simulations. Ring penetrations is a prevalent problem that leads to unrealistic physical behavior in simulations, and LongBondEliminator provides a powerful solution and has the potential to enhance the accuracy of simulations for a number of applications.
The problem of ring penetrations
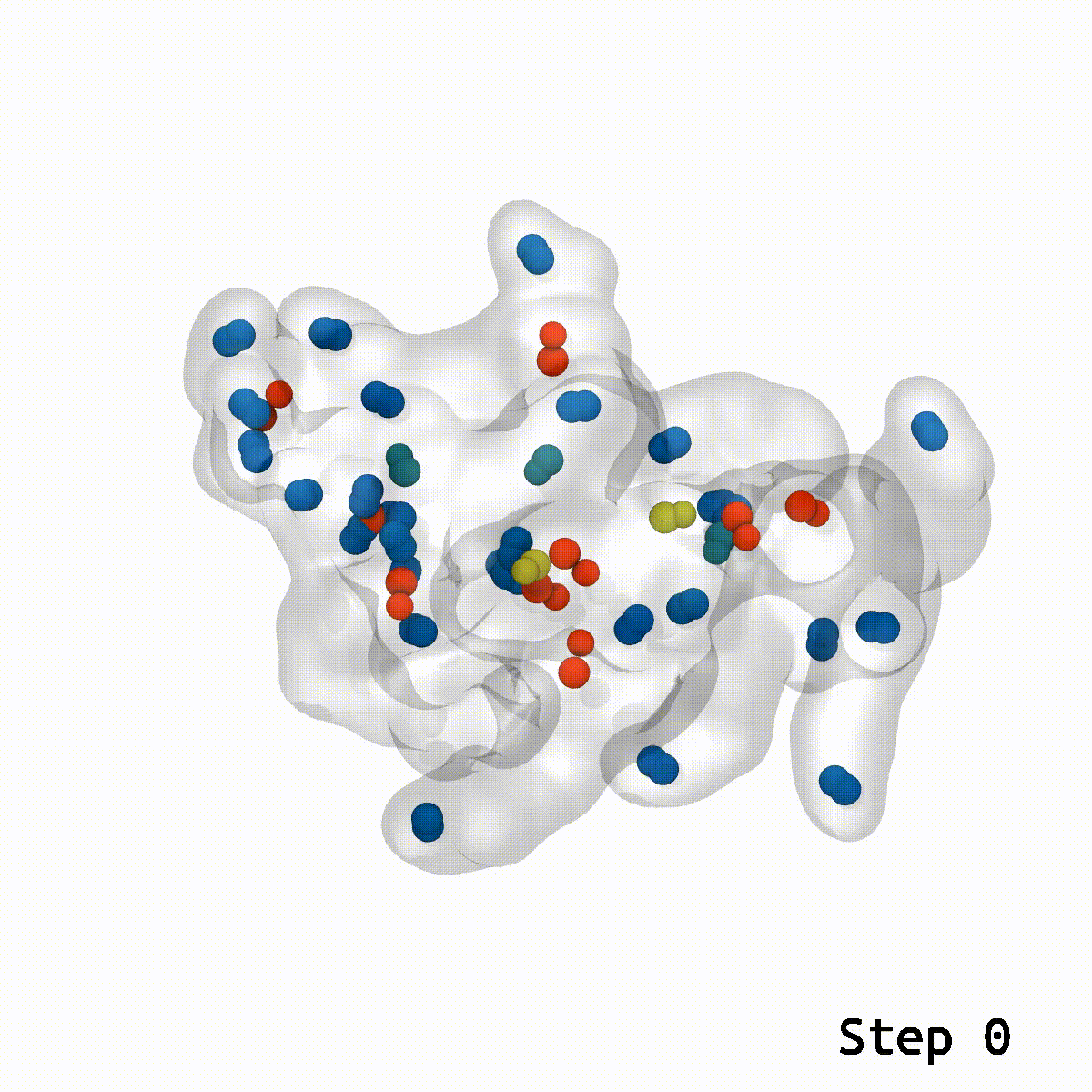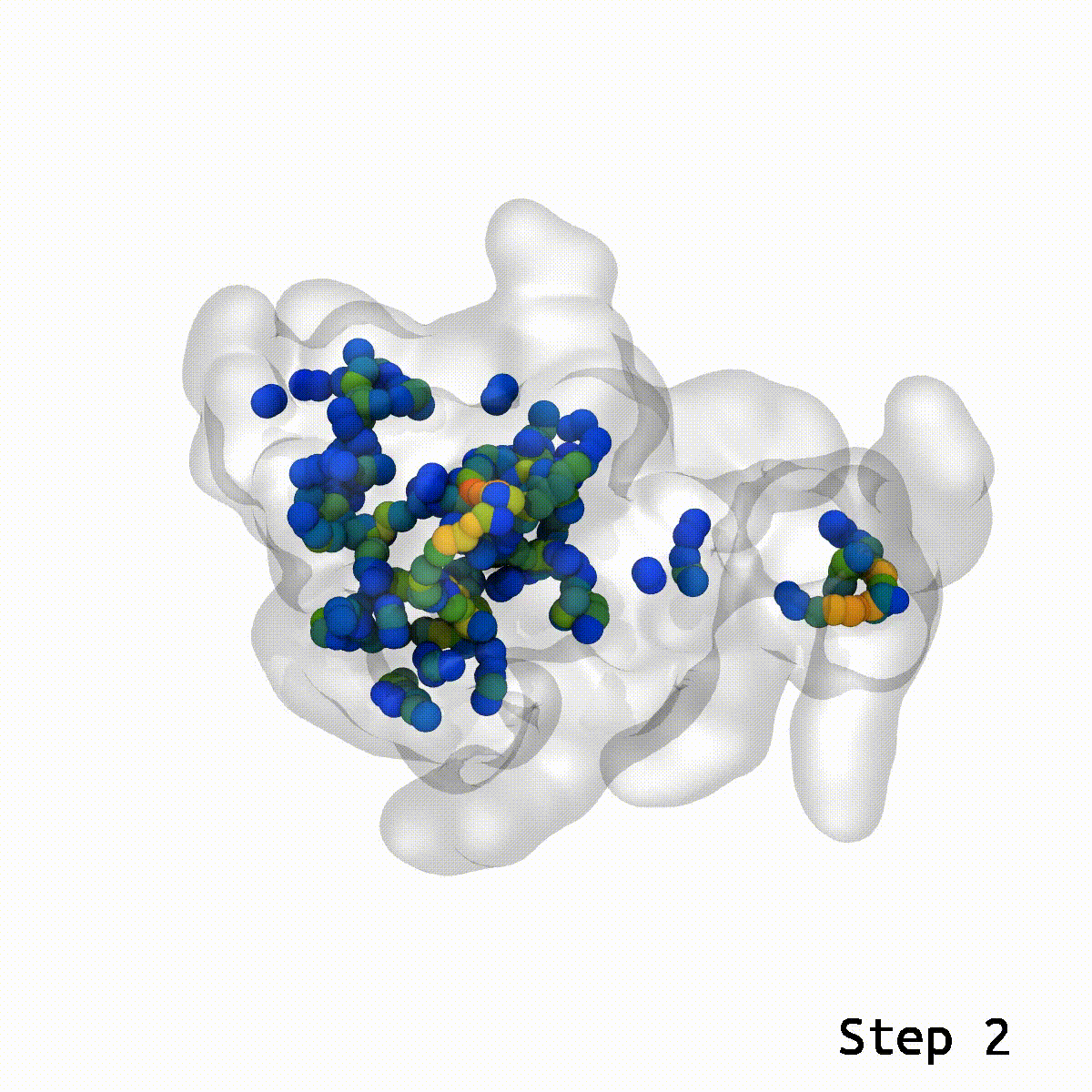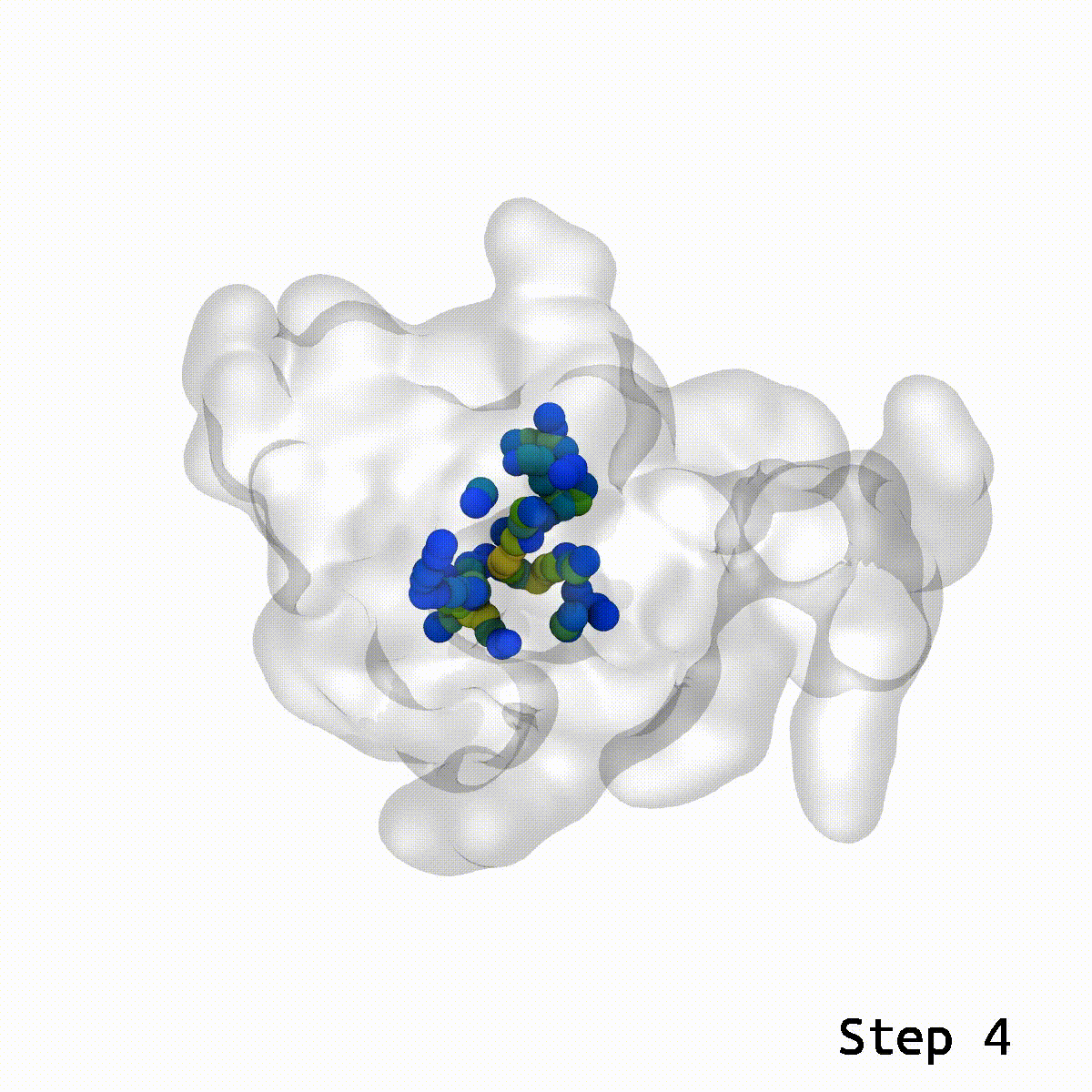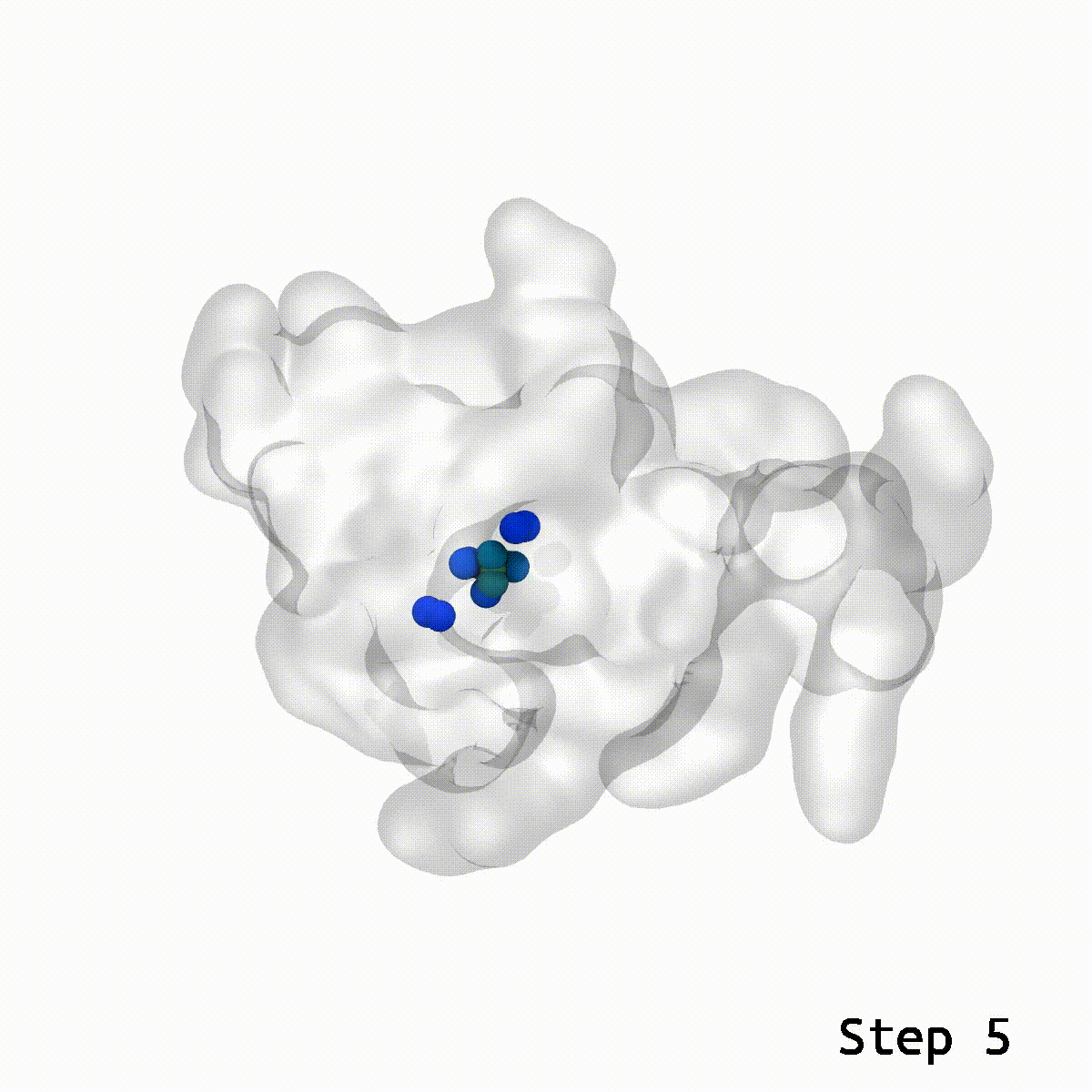
Ring penetrations occur when two or more molecules overlap, which leads to unrealistic and unphysical behavior in the simulations of a molecule. LongBondEliminator is a tool designed to detect and fix ring penetrations in simulation systems and can be used as a plugin for the molecular visualization program Visual Molecular Dynamics (VMD). This study provides three examples of the tool’s utility in the elimination of ring penetrations in increasingly complex systems: a lignin polymer, a viral capsid assembly, and a highly glycosylated protein aggrecan.
Accurate simulation of complex biological systems in molecular simulation is a challenging problem, particularly with regard to the precise positioning of multiple components in crowded environments. While structural databases and AI-powered prediction approaches have made this process easier for basic systems, adding new parts might result in difficult-to-minimize nonphysical structures. Existing methods, such as human modifications or environment-aware algorithms, may not scale effectively with growing complexity or size, necessitating the development of new tools.
Ring-piercing artifacts are a prevalent problem in molecular modeling, especially when newly inserted entities cause two nominally coupled atoms to be on opposing sides of a small ring. This can result in awkward non-physical arrangements independent of the simulation technique. With quantum mechanical or machine learning techniques, the bond will be divided throughout the ring, generating an alternate topology, whereas the pierced ring configuration represents a local minimum in classical systems. While the traditional strategy of treating the entire system alchemically and minimizing may repair many systems with modest atomic overlap, more robust methods may sometimes be required in a few cases. Many toolkits, such as CHARMM-GUI and current toolkits for mixing lipids, include methods for identifying ring penetrations and can automatically resolve some ring penetrations produced by sterols.
LongBondEliminator
LongBondEliminator (LBE) is a plugin for the molecular simulation engine NAMD that can identify and correct ring-piercing errors caused by the addition of new entities to the system. The instrument utilizes a module for collective variables, density-based forces, and alchemical procedures to break the lengthy link. The researchers illustrate the efficacy of LBE with three instances of increasing complexity: a lignin polymer, a viral capsid assembly, and a bulky, highly glycosylated protein. The application is accessible via gitlab and Zenodo.
Working of LongBondEliminator
The LongBondEliminator tool was created empirically to eliminate ring-piercing artifacts found when constructing lignin polymers. The LBE instrument identifies long, stretched bonds in a given minimal structure, which are indicative of ring-piercing artifacts. The software then uses a mix of approaches, including alchemical free energy perturbation (FEP) methods, GridForces, and the collective variables (colvars) module inside NAMD, to break the lengthy link and force overlapping atoms apart. The initial distance cutoff of 1.65 Å was somewhat larger than the equilibrium bond length within lignin; however, LBE now employs a variable distance cutoff based on the equilibrium bond lengths obtained in the simulation parameter files for the CHARMM force field. The minimization steps will continue until the longest bond in the system, when reduced without flagged alchemical atoms, is less than 0.4 Å longer than the bond length at equilibrium. The run will end after 100 minimization stages of 500 steps each to avoid endless loops. LBE is an all-purpose tool that can handle a broad variety of ring-piercing artifacts with various beginning configurations.
If stretched bonds reach 0.4 Å during minimization, LigninBuilder provides many biassing methods to resolve them. These involve creating a repulsive density and collective variable around the lengthy connection and alchemically manipulating nearby atoms. These methods may not work for complex glycosylation situations. To fix this, LigninBuilder introduced additional workflows. The program now prioritizes weak links and selects lengthy ties within 0.1 Å of the system’s most stretched bonds. The algorithm now solves long connections repeatedly, which stabilizes complicated piercing artifacts. LigninBuilder also uses a collective variable to shift the two atoms perpendicular to the bond vector and multiplies the force constants by two if the same bond survives across iterations. LigninBuilder limits all chiral centers in the molecule using NAMD’s extrabonds function to minimize excessive stresses on individual atoms. These devices correct ring piercings with minimum user input.
Three examples have been provided in the paper to illustrate the efficiency of LigninBuilder’s LongBondEliminator (LBE) in resolving ring-piercing artifacts in various systems. The three examples are a tiny lignin polymer, the viral envelope of the AstraZeneca COVID-19 vaccine, and aggrecan, a heavily glycosylated protein. The researchers minimized computational cost by using NAMD 2.14 without hydrating water molecules and by providing a Tcl script for repeated minimizations in VMD. As they worked in a vacuum, the authors utilized a dielectric of 80 and did not employ PME for long-range electrostatics.
Test Cases
Lignin Builder
For the first example, a lignin polymer with 5322 atoms was used, which is quite big for lignin. Several ring penetrations were constructed with LigninBuilder for the original construction. The system was reduced using the CHARMM force field for lignin with 12 Å nonbonded cutoffs.
Virus
LBE is a method utilized to avoid collisions in big biomolecular systems, notably in viral capsid assemblies, which are susceptible to significant atom clashes and ring piercings due to rigid body transformation-based docking methods. LBE makes use of the CHARMM36m force field to reduce interactions between protein components and enhance docking techniques. LBE was applied as a test case to resolve ring penetration artifacts in a recently disclosed viral capsid structure. When the LBE procedure concludes, cryo-electron microscopy data can be refined further.
Glycosylated protein
In this study, LBE was utilized to enhance the structure of aggrecan, a heavily glycosylated protein. Using simulations of directed molecular dynamics and equilibrium molecular dynamics, the original structure was stretched and then relaxed. Using a VMD script, the carbohydrate chains were automatically linked to the relaxed core protein structure. Due to the random rotation of amino acid side chains, however, the carbohydrate chains were entangled with one other and the protein backbone, resulting in nearly 800 hexose ring perforations. Using the CHARMM36m force field for proteins and sugars and 12 nonbonded cutoffs, LBE was then utilized to reduce the structure and resolve these piercings.
Analysis
Using various tools, the geometry of the intermediate and final structures in the test instances was examined. The longest bond after each minimization step was recorded using “getlargestdeviation,” and a depiction of the degree of bond extension for individual atoms was created using “tagdeviation.” In the viral capsid test case, MolProbity and clashscores were monitored during the minimization, and PHENIX was used to examine the intermediate structures. Using Python packages such as NumPy and Matplotlib, the findings were plotted.
Results of the test cases
LongBondEliminator focuses on the degree to which a bond is stretched and employs this as its primary measure of convergence. The length of the longest link throughout multiple workflow cycles was displayed. This graph demonstrated that only 20 cycles are necessary to achieve a structure with bond stretches less than 0.4 for the selected test situations. The soft-core potential permits the stretched bond to relax and approach its equilibrium length, and it may even go below the cutoff. Yet, the visual models revealed that there are still mildly stretched bonds in the final state, as seen by the blue spheres whose bonds are still stretched by more than 0.1. At this degree of extension, the bonds can be utilized as inputs for actual simulations. LBE targets the longest bonds first, and it is possible that some early ring penetrations are not initially addressed. These new longest bonds are only targeted in consecutive phases until every stretch or deviation of the bonds falls below the selected threshold.
LongBondEliminator conducts alchemical transformations to possibly deficient beginning structures, and it is crucial to monitor the quality of intermediate structures at each stage. A plot between the clash and MolProbity scores for various asymmetric units in a facet of an adenovirus with T25 point symmetry revealed that the original structure constructed from many subcomponents might contain hidden clashes. Due to ring penetration artifacts in the original structure, the clash and MolProbity scores increase in step 1 following unperturbed reduction. Yet, additional LBE processes lessen atomic clashes within the structure and enhance the overall quality of the structure. The MolProbity scores may be viewed as resolution-equivalents, and the lower the value, the greater the quality of the structure.
Conclusion
The LBE process is not founded on completely new concepts but rather merges existing concepts into a single instrument. GROMACS users are advised, for instance, to employ alchemical transformations to address defective atomic contacts. Soft-core potentials are frequently utilized to lower the expense of bringing two atoms together. Moreover, biases utilizing collective variables or density grids are used to direct systems down productive paths. LongBondEliminator is innovative since it combines these principles into a single tool that can be readily integrated into VMD structure construction operations.
Article Source: Reference Paper
Learn More:
Sejal is a consulting scientific writing intern at CBIRT. She is an undergraduate student of the Department of Biotechnology at the Indian Institute of Technology, Kharagpur. She is an avid reader, and her logical and analytical skills are an asset to any research organization.