Large language models, such as GPT (Generative Pre-trained Transformer), have showcased remarkable proficiency in various natural language processing tasks. Their ability to understand and generate text akin to human speech has paved the way for their widespread application across diverse domains. However, integrating these language models into specialized disciplines like biology has been challenging. This article introduces Cell2Sentence (C2S), a novel method designed to adapt language models to the intricacies of biological data, particularly focusing on single-cell transcriptomics.
Cell2Sentence has vast potential for applications, including generating rare cell types, identifying gene markers, and analyzing transcriptomics data using natural language. Models have been developed to comprehend and generate transcriptomics data, interacting in a human-like manner by leveraging the extensive pretraining of Large Language Models (LLMs). This approach is noteworthy for its flexibility and simplicity, aligning with industry-standard language modeling training pipelines and making it accessible to consumers through well-established third-party libraries.
The data transformation pipeline, available on GitHub, seamlessly integrates preprocessing unprocessed single-cell data into the C2S format and postprocessing generated texts back into gene expression vectors. This ease of use empowers users to customize the training process, adding metadata or designing personalized prompts to suit their specific requirements.
While Large Language Models (LLMs) like GPT have demonstrated outstanding capabilities in natural language processing tasks, their effective integration into specialized domains such as biology remains a significant hurdle. Single-cell transcriptomics, a critical field for studying individual cells, poses a unique challenge for leveraging LLMs. Existing techniques often overlook the potential of pretrained LLMs, relying heavily on specialized neural networks.
In natural language processing, LLMs have excelled in tasks like text classification (e.g., BERT), question answering (e.g., LlaMA-2), and text generation (e.g., GPT-3). Conversely, in single-cell transcriptomics, specialized deep neural networks are employed for cellular annotation, batch effect removal, and data imputation, drawing from datasets like GEO and HCA. Efforts are underway to establish foundational models akin to NLP, with advancements like prompting (e.g., GPT-2) and multimodal training (like C2S) propelling these fields. C2S stands out by directly transforming data into text for embedding, a novel approach in single-cell transcriptomics.
Cell2Sentence: Adapting Language Models to Biology
Cell2Sentence allows meaningful manipulation of cells as text demonstrated through training LLMs on cell sentences. Larger models achieve lower perplexities, indicating the capacity of causal language models to comprehend cell sentence semantic distribution from a narrow single-cell RNA-seq dataset.
Assessing cell reconstruction, the models show promising results in generating realistic cells, which is crucial for downstream analyses. Generated cells from fine-tuned models exhibit a high correlation with real cells across different cell types, capturing a significant portion of the expression variation. Notably, initializing a model with a pretrained language model significantly improves performance, emphasizing the importance of mutual information for understanding cell sentence generation.
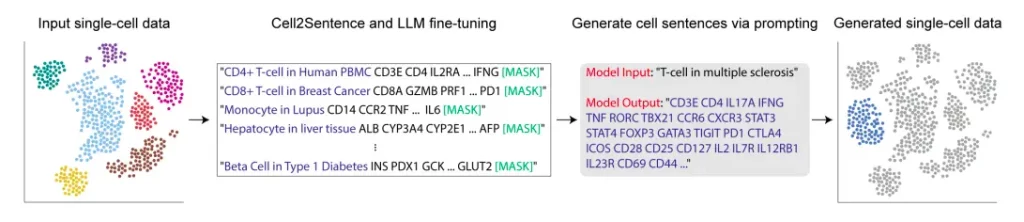
Image Source: https://doi.org/10.1101/2023.09.11.557287
The Cell2Sentence approach revolves around representing gene expression data, a fundamental component of single-cell transcriptomics, as textual information. Each cell’s gene expression profile is transformed into a sequence of gene names arranged by their respective expression levels. This sequence, which we term “cell sentences,” acts as a unique textual representation of a given cell.
To further fine-tune these language models for biological understanding, they utilize causal language models like GPT-2. This process involves training the model on cell sentences, where the language model learns to generate biologically valid cells when provided with a cell type as a prompt. Conversely, it also learns to predict cell type labels accurately when given cell sentences as input.
Enhancing Model Performance
The important finding is that these language models perform significantly better on difficulties involving cell sentences when they have been pre-trained on problems requiring normal language. The language model gains a deeper comprehension of biological data, enabling it to generate physiologically plausible cells and make accurate predictions about the types of cells.
Each cell’s gene expression profile is transformed by C2S into a textual list of gene names sorted according to expression level. Any pretrained causal language model can be adjusted for these cell sequences thanks to this conversion. Notably, using C2S alone dramatically degrades model performance compared to adding natural language pretraining followed by C2S training, with performance scaling with model size. The improved models show the capacity to produce cells based on gene name sequences, produce cells using natural language prompts, and generate natural language descriptions about cells.
Generating Biologically Valid Cells
When prompted with a specific cell type, the fine-tuned GPT-2 model generates biologically valid cells, showcasing its ability to understand and produce meaningful biological information. This is a testament to the adaptability and versatility of large language models in specialized domains like biology.
Accurate Cell Type Label Prediction
Conversely, the fine-tuned GPT-2 model accurately predicts cell type labels when given cell sentences as input. This demonstrates the bidirectional applicability of our Cell2Sentence approach: the model can not only generate biologically valid information but also interpret and classify cells based on their gene expression profiles.
Cell Sentence Encoding: A Reliable, Reversible Transformation
Although genes are not intrinsically ordered in transcript matrices, their expression patterns adhere to inverse-rank frequency patterns, establishing a consistent relationship between a gene’s expression level within a cell and its rank among the expressed genes in that cell. This inverse-rank relationship is modeled using a log-linear distribution, approximated in log-log space through linear regression. This modeling allows for conversion between gene rank and expression domains, forming the basis for generating rank-ordered sequences of gene names used to train subsequent language models.
C2S has been proven viable in both forward and turnaround changes with negligible data loss. The assessment shows that the change to cell sentences and back to expression space saves the fundamental data in single-cell information, empowering examination in the normal language space of cell sentences and precise transformation back to expression.
Opening Potential: A Combination of Language and Science
The Cell2Sentence approach addresses a critical headway in overcoming any barrier between regular language understanding and natural comprehension. Consolidating the force of existing language models with the language of biology makes for a versatile system for investigating and deciphering single-cell transcriptomics information.
The Final Word: Unveiling Insights
The innovative approach of “Cell2Sentence” encapsulates gene expression data into easily digestible sentences for language models, markedly enhancing model performance on cell-related tasks compared to models trained from scratch or specialized for single-cell RNA sequencing data. This blending of language models and biology opens up new avenues for investigation, evaluation, and application in the life sciences, expanding our knowledge of biological systems in the process. This integration allows seamless incorporation of cell sentences with textual annotations, enabling effective generation and summarization tasks, all benefiting from pre-training on natural language.
However, there are certain acknowledged limitations. Cell2Sentence represents gene expression data by sorting gene names based on expression levels, losing specific quantitative information about expression levels. The model must infer expression strength solely from gene order.
However, current research is restricted to a few distinct cell types and functions. To prove the versatility and dependability of Cell2Sentence, it is essential to adapt it to various datasets and investigate a wider range of applications utilizing current natural language model architectures. This research establishes the groundwork for fusing computational biology with language models, opening up a world of exciting possibilities.
Story Source: Reference Paper
Learn More:
Prachi is an enthusiastic M.Tech Biotechnology student with a strong passion for merging technology and biology. This journey has propelled her into the captivating realm of Bioinformatics. She aspires to integrate her engineering prowess with a profound interest in biotechnology, aiming to connect academic and real-world knowledge in the field of Bioinformatics.