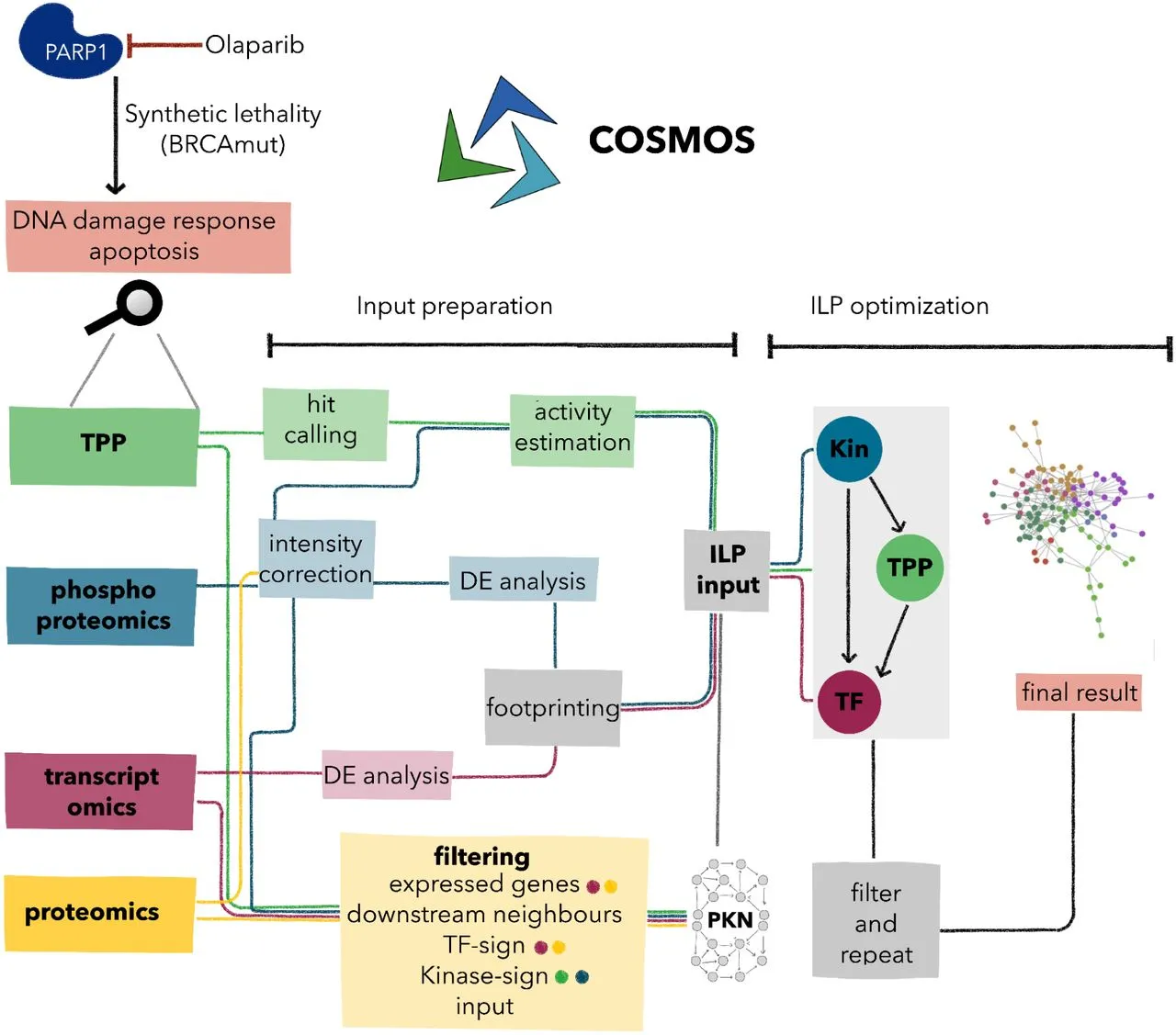
Understanding diverse molecular mechanisms is necessary for the research of complex diseases. Using COSMOS, the integration of Thermal Proteome Profiling (TPP) with phosphoproteomics and transcriptomics provides a comprehensive perspective of biological activities. The technique emphasized the importance of TPP in functional proteomics by discovering both well-known and novel molecular responses to Poly (ADP-ribose) polymerases (PARPs) inhibition in ovarian cancer cells. By combining a variety of omics data, integrated network models highlighted the potential of TPP integration to expand knowledge of complex biological systems across numerous disorders.
Complex changes in protein function and abundance occur throughout cellular regulation. Phosphoproteomics demonstrates protein function and kinase activity, whereas Thermal Proteome Profiling (TPP) tracks changes in protein thermal stability. Transcriptomics is the study of transcription factor activity. TPP and phosphoproteomics integration increases comprehension by highlighting functionally relevant alterations and associated signaling linkages. COSMOS, a computer integration method, uses biological networks to infer enzyme activity and causal routes. Using COSMOS to investigate PARP inhibition and DNA damage revealed functional alterations, enhancing our understanding of drug response mechanisms. This integrated strategy has the potential to improve cancer treatment options.
Multi-omic Profiling of Olaparib Response in Ovarian Cancer Cells
In investigating Olaparib’s effect on UWB1.289 cells, it employed transcriptomics, phosphoproteomics, and TPP. Transcriptomics revealed 44 significantly altered genes among 20,493 expressed genes. Phosphoproteomics identified 256 significantly changed phosphosites among 11,615, with emphasis on the ATM-ATR axis and downstream CDKs. TPP data identified 76 proteins with altered thermal stability in response to Olaparib, indicative of diverse molecular effects such as drug binding or complex formation.
Footprint Analyses for Molecular Insights:
This helps to estimate kinase and transcription factor activities based on phosphoproteomics and transcriptomics data. The top 30 activated transcription factors (TFs) were primarily associated with interferon, nuclear receptor, DNA repair, and cell cycle signaling.
In the phosphoproteomics data, deregulated kinases were notably linked to the ATM-ATR axis and downstream CDKs, aligning with expected cell cycle arrest induced by PARP inhibitors. This showed the complementary molecular insights gained from transcriptomics, phosphoproteomics, and TPP, providing a comprehensive understanding of the cellular response to Olaparib.
Integration via COSMOS Connects Data Modalities:
COSMOS was used to integrate transcriptomics, phosphoproteomics, and TPP data, leveraging prior knowledge to create causal molecular networks. Incorporating a few transcription factors, kinases, and TPP measurements, various approaches were applied, and optimal results were observed by utilizing kinases as upstream regulators, TPP proteins as intermediates, and transcription factors as downstream effectors. This approach produced a robust and reproducible molecular network, effectively connecting information from the three omics layers.
Enhanced Biological Insights with TPP Inclusion:
Incorporating TPP data in the integrated networks significantly enriched biological information, especially in pathways related to cell cycle and DNA damage response (DDR). The clustering and enrichment analysis highlighted the unique value of TPP data in unraveling processes like interferon signaling and YAP/TEAD signaling. Further pathway-specific analyses demonstrated the critical role of TPP proteins in signaling cascades, showcasing their involvement in complex biological processes, including DNA damage checkpoint and cell cycle regulation, and highlighting potential phosphorylation-induced stabilization effects.
Interferon Signaling and YAP-TEAD Signaling Investigations:
The study explored the lesser-known aspects of PARP inhibition by delving into interferon signaling, a reaction acknowledged but less explored. Key transcription factors like RELA, STAT1, STAT2, STAT3, and IRF1 were found to orchestrate this response, supported by TPP hits MX1 and OAS1, activated through interferon signaling. The thermal stabilization observed aligned with transcriptional shifts triggered by this stimulus, shedding light on this intriguing molecular response.
Integrating Omics Data for Insights into PARP Inhibition Response:
Integrating multiple omics data, including Thermal Proteome Profiling (TPP), phosphoproteomics, and transcriptomics through COSMOS, revealed the comprehensive molecular response to Olaparib in UWB1.289 cells. Each omic profile offered unique insights, and the integration enriched the understanding of PARP inhibition effects. TPP, with its functional focus, emerged as a valuable tool in multi-omics research, presenting a pathway to refine modeling and broaden insights into the intricate dynamics of drug-induced molecular responses, promising a deeper exploration of complex molecular dynamics in drug-related contexts.
Experimental Procedures for Multi-Omics Profiling
UWB1.289 cells (a type of cell) were cultivated in a controlled environment with the proper nutrients. After this, cells were treated with varied amounts of Olaparib or control to examine the drug’s effects. The cells were extracted and processed for analysis after treatment.
To understand genetic activity, RNA was collected and sequenced for transcriptomics. Proteins were isolated, processed, and analyzed for proteomics and phosphoproteomics to understand protein behavior and change better. In addition, Thermal Proteome Profiling was performed to investigate how protein structures change with temperature. These in-depth studies were critical for understanding the cellular response to Olaparib. Mass Spectrometry (MS) was utilized for relative quantification in proteomics, providing a comprehensive multi-omics view and shedding light on the intricate molecular dynamics in drug-induced DNA damage responses.
Data Analysis
To gain insight into how cells respond to Olaparib, a possible anti-cancer therapy, genetic and protein data were analyzed. For the genetic portion, raw sequencing data (FastQ files) was processed to provide gene activity counts. The counts were then mapped to the human genome, duplicate sequences were discovered and counted, and readings were given to individual genes. Genes with sufficient numbers were chosen for further investigation. Statistical methods were used to determine which genes’ activity changed significantly following Olaparib treatment.
Protein modifications (phosphorylation) were maintained in focus for the protein section. For appropriate comparison, the intensity of these changes was analyzed and normalized. To ensure dependability, data was filtered to include proteins having sufficient information for analysis. Similar to genetic analysis, statistical methods were used to identify proteins with significant changes in their modification levels due to the treatment.
A computational technique was also used to estimate the activity of specific biological entities (transcription factors, kinases, and phosphatases). To understand how these entities influence the cellular response to Olaparib, information from known relationships in biological networks was incorporated.
Thermal Proteome Profiling (TPP) is a specialized analysis that examines how protein stability changes with temperature variations in order to identify proteins that responded strongly to Olaparib treatment in terms of stability.
Pathway analysis involves grouping related genes or proteins to examine the results of protein activity analysis. This allows researchers to gain insights into the specific pathways linked to medication response. Finally, clusters of biological processes were grouped and compared to gain a better understanding of the cellular changes generated by Olaparib. This intricate analysis allowed to unravel the complex biological responses triggered by this potential anti-cancer drug.
In summary, TPP is integrated with phosphoproteomic and transcriptomic data via COSMOS, providing a holistic understanding of the molecular response to PARP inhibition. This approach showcases the potential for integrated functional proteomics to study complex molecular processes comprehensively.
Article source: Reference Paper
Important Note: bioRiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Prachi is an enthusiastic M.Tech Biotechnology student with a strong passion for merging technology and biology. This journey has propelled her into the captivating realm of Bioinformatics. She aspires to integrate her engineering prowess with a profound interest in biotechnology, aiming to connect academic and real-world knowledge in the field of Bioinformatics.