Scientists from Princeton University have developed a comprehensive model of the structure and function of the open reading frame (ORF) 1 of Hepatitis E Virus (HEV), a type of RNA Virus that causes more than 20 million infections yearly. The ORF1 is a multifunctional protein essential for viral replication, however, it is still being determined how its subunits interact. Studies show that ORF1 is not subject to proteolytic processing. The model discussed in this article sheds light on the vital role of interdomain interactions in viral replications.
Problems in finding HEV therapy
Hepatitis E virus (HEV) is a significant threat to world health, particularly among immunocompromised persons and pregnant women, the latter of which have a mortality rate of nearly 30 percent in the third trimester. There is currently no therapy approved for HEV, and the available medicines have substantial limits and adverse effects.
HEV is a (+) ssRNA virus whose genome has three partially overlapping open reading frames (ORFs). ORF1 encodes the viral replicase and has seven domains, of which only four have been functionally defined. Regarding its function and targets, the putative papain-like cysteine protease (pPCP) domain in ORF1 has been the topic of controversy, with data both supporting and disputing its activity. Yet, it remains possible that this region hosts other activities. A better knowledge of the HEV replication cycle and the role of ORF1 might facilitate the development of effective HEV treatments.
Recent studies have revealed incomplete structural information on tiny areas inside ORF1, but these regions are not functional outside of the ORF1 protein. To overcome this limitation, scientists studied the ORF1 protein using biochemical, genetic, mass spectrometric, and computational methods.
Role of protein structure and metal ion binding
It was shown using a trans-complementation technique that the pPCP domain is exclusively functional within the context of the full-length ORF1 protein. The authors generated testable predictions regarding the ORF1 protein structure using the AlphaFold method, a recent advance in protein structure prediction. They discovered that the loss of replicative capability induced by site-directed mutagenesis was not related to a lack of proteolytic activity but rather to a loss of structural integrity resulting from an inability to bind divalent metal ions.
Furthermore, the authors purified the ORF1 protein for subsequent inductively coupled plasma mass spectrometry (ICP-MS) study using a tolerable epitope locus within the hypervariable region of ORF1. They discovered that point mutations in either the pPCP or Y-domain bound to divalent metal ions with different capacities. This shows that HEV ORF1 acts as a single big multidomain protein that does not undergo proteolytic processing and that the putative catalytic residues indicated by previous bioinformatic investigations are structural in nature due to their capacity to bind divalent metal ions.
Catalytic residues in HEV replication
Based on bioinformatics analysis, the functional domains and genomic organization of the Hepatitis E Virus (HEV) were originally hypothesized in 1992. Based on low sequence identity with the distantly related rubella virus, the putative protease domain in HEV ORF1 was derived. The production of infectious clones of cell-culture-suited strains and reporter replicons employing GFP or Gluc, as well as other tools, have been created to examine the HEV genome systematically. C483 and H590 residues were altered to chemically comparable amino acids to test the significance of the hypothesized catalytic duo. The results demonstrated that genomes with mutations at locations C483 and/or Y590 were incapable of sustaining sustained replication. The results indicate that the hypothesized catalytic duo is essential for effective RdRp-mediated viral replication.
Next, it was determined if the absence of viral replication seen after altering the highly conserved C483 residue in the Kc1/p6 cell culture-adapted strain of HEV was also detected in other known human-tropic HEV strains. In SAR55, SHEV3, and TW6196E genomes carrying the C483A mutation, the replication of HEV RNA was greatly impeded. Encoding the alternate cysteine had a little negative effect on viral replication efficiency, however, the use of any alanine codon reduced viral replication to the level of the Pol (-) mutant. This indicates that the requirement of the C483 residue lies in protein folding or function, despite the fact that Y590 is variable among HEV virus genotypes.
Transcomplementation
To analyze the mechanism behind the functional defects of the C483 mutants, the researchers utilized a previously established transcomplementation strategy to evaluate the role of HEV ORF1 in viral genome replication. In HepG2C3A cells, they produced wild-type ORF1 or mutant forms of ORF1 and transfected in vitro-transcribed RNA from the Kernow C1/p6 Gluc wild-type, Pol (-), or C483A genome. The results indicated that the mutant version of ORF1 had a detrimental effect on the replication of the wild-type replicon, most likely due to the mutant protein’s competitive inhibition of the replicon RNA.
However, the production of wild-type ORF1 was able to restore replication defects caused by the Pol (-) or C483A mutations, indicating that the activities of this region of ORF1 are necessary for viral replication. This transcomplementation method allows for the independent study of the activities of particular sections of ORF1, such as polyprotein processing and host cellular environment regulation, independent of viral genome replication.
Investigation of essential elements for HEV replication using AlphaFold
The researchers analyzed the pPCP sequences of all eight HEV genotypes and identified an octa-cysteine motif. They then utilized mutagenesis on the Kc1/p6 strain of HEV to establish the significance of each cysteine to viral replication. They discovered that just the core six cysteines were required for replication, while the first and final cysteines were either redundant or mildly harmful. The researchers next ran an unbiased genetic mutagenesis screen of the whole pPCP region and discovered a number of triplicates that were essential for viral replicative fitness. It was shown that conserved cysteines and a variable amino acid at position 590 are essential for viral replication, although some lengths of amino acids downstream of the hexa-cysteine motif are mutagenesis-tolerant.
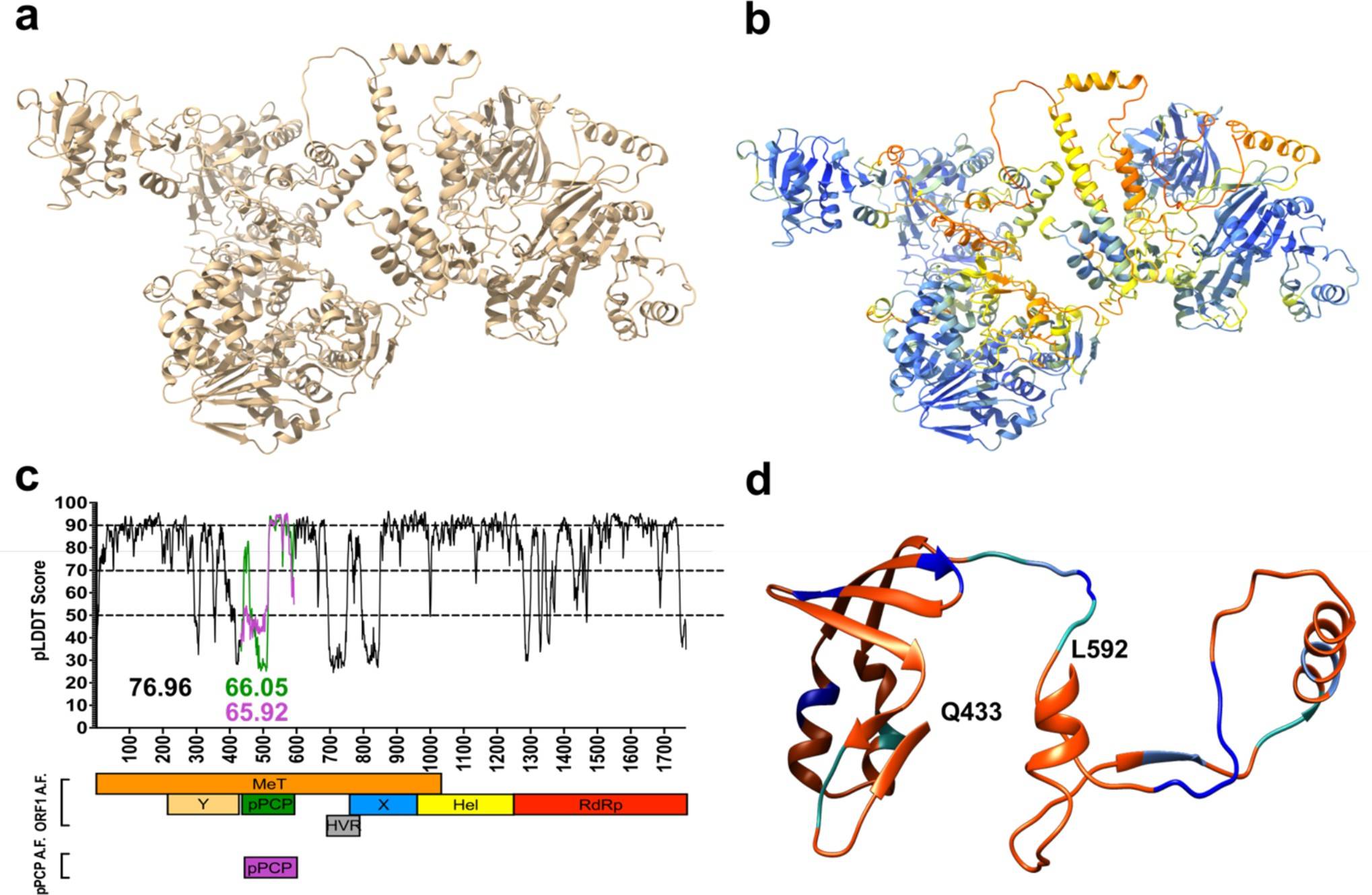
The researchers used AlphaFold to estimate the structure of the HEV ORF1 protein and examined the prediction’s confidence levels. They also assessed the prediction’s accuracy by comparing it to the resolved structures of HEV ORF1 segments and macro domains of distantly related viruses. Despite having a low sequence similarity, they discovered a significant degree of local alignment identity between the predicted structure and the solved structures. This indicates that the predicted structure is likely accurate and can provide light on the domain organization and protein folding of HEV ORF1.
AlphaFold was able to predict the secondary structures of known sections of HEV ORF1 with high confidence but was unable to predict the structure of the disordered area. This indicates that the disordered area is unlikely to fold into a protease domain, which was validated by evaluating the results of alanine-scanning mutagenesis.
Impact of Mutations on Metal Ion Binding and Replication
The researchers used AlphaFold to analyze the predicted folding structures of wild-type ORF1 and two mutants (C483A and C563A) to examine the implications of mutations within the pPCP domain on HEV replication. They discovered that C483A is completely replication deficient, whereas C563A is replication blunted. Researchers discovered a unique pseudo-zinc-finger produced by the amino acids beneath the HEV hexacysteine motif, which is damaged by the C483A mutation. They also discovered that the mutations impair the divalent metal ion-binding domains within the pPCP, causing structural domains essential to viral replication to develop abnormally and preventing HEV ORF1 from replicating effectively.
Next, the metal ion-binding activity of the ORF1 protein in the Hepatitis E virus was examined (HEV). Prior research had indicated that some sections of ORF1 would include calcium and zinc ion binding sites. Until recently, it had been challenging to purify a replication-competent ORF1 protein due to a paucity of well-characterized ORF1-specific antibodies. The study examined the impact of mutations on ORF1’s capacity to bind divalent ions using a HA-tagged ORF1 protein overexpression system. The data show that divalent ion binding ability impacts appropriate structural confirmation of ORF1, resulting in varied replication capacity dependent on mutation location.
Subcellular Localization and Replication
The study used confocal microscopy to investigate the subcellular localization patterns of various ORF1 mutants. The findings demonstrated that mutations in the pPCP and upstream Y-domain impact ORF1’s ability to localize to the nucleus and form aggregates in the cytoplasm, which corresponds with their replicative capability. Replication-deficient mutants, such as C483A-HA and H249A-HA, lost their capacity to localize to the nucleus, whereas replication-competent mutants, such as C563A-HA and D248A-HA, retained their ability to establish replication and share localization patterns with ORF1 WT-HA. These findings suggest a potentially novel mechanism by which mutations in these domains interfere with HEV’s replication cycle.
Conclusion
The study discusses the use of reporter replicons for studying the functional domains of the HEV replicase, with a focus on the pPCP domain of ORF1. The study found that mutations in the core cysteine residues of pPCP render HEV replication incompetent and that the dysfunction is likely at the protein level. The paper also states that the roles of pPCP are still contested and that HEV ORF1 is likely a big multidomain protein. The potential that unexplored domains exhibit cis-acting processing activity remains an intriguing topic of discussion.
The researchers conducted a mutagenesis screen to determine which motifs within the pPCP domain of the HEV virus were essential and which were not. They discovered that the HEV motif is structurally significant and likely plays a role in metal ion binding. They employed AlphaFold to get structural insights and discovered that mutations in conserved cysteines or histidine affected the assembly of zinc fingers and lowered ORF1 localization. By purifying ORF1 protein and exposing it to ICP-MS and confocal microscopy, they validated their expectations. This work proved the effectiveness of combining AI-driven predictions with hypothesis creation and testing to uncover functional areas within ORF1.
Article Source: Reference Paper
Learn More:
Sejal is a consulting scientific writing intern at CBIRT. She is an undergraduate student of the Department of Biotechnology at the Indian Institute of Technology, Kharagpur. She is an avid reader, and her logical and analytical skills are an asset to any research organization.