Scientists from Stanford and Harvard Universities have developed a groundbreaking genomic foundation model called HyenaDNA, which promises to transform the analysis of DNA sequences. The model addresses the limitations of previous approaches by offering the handling of long sequences (up to 1 million tokens), single nucleotide resolution, higher quality results, and reduced time complexity compared to attention-based models.
For a very long time, biologists and deep learning researchers have worked to comprehend and learn crucial knowledge from genomic sequences about gene regulation, protein synthesis, and various other cellular processes. To achieve their goal of decoding genomic sequences, scientists utilized foundation models in genomics. Machine learning models called foundation models are pre-trained on a lot of data and discover the broad patterns of the input data.
Previous Models Have Had Success with Proteins, so Why Not with DNA?
Scientists envision the possibility of mapping DNA instructions (genotypes) to observable traits (phenotypes). Deciphering the information embedded within DNA sequences can significantly help in predicting the locations and functions of genes, recognizing regulatory elements (promoters, enhancers, etc), and even examining the evolution of species. In the case of proteins, the mapping of instructions to observable traits has been successful with already existing language models, but the same is not true for DNA because the length of DNA sequences belongs to orders of magnitude much higher than proteins. The human genome has 3.2 billion nucleotides in length. Also, DNA has long-range interactions among its constituents that span over 1,00,000 nucleotides.
The previous models were dependent on attention-based transformers, and attention-based mechanisms scale quadratically, meaning that the increase in the sequence length is proportional to the square of the computational cost of attending to all positions in the sequence. Due to quadratic scaling, the models could pre-train only on tokens of sizes 512 to 4,096, which is even less than 0.001% of the entire human genome. The models also relied on fixed K-mers or tokenizers to group DNA into meaningful units of a specific length, similar to words in natural language that fail to capture minute details, like the changes in single nucleotides that can happen in single nucleotide polymorphisms (SNPs), which profoundly influence the regulation and expression of genes. Thus, long-range context and single nucleotide resolution are critical in efficient genome processing.
HyenaDNA: Algorithms and Performance for Long-Range Genomic Sequence Processing
To combat the drawbacks of attention-based transformers, scientists propose applying foundation models that can learn general patterns from unlabeled genomic data and optimize them for downstream processes such as identifying regulatory elements. Recently, Hyena, a language model based on implicit convolutions (the model does not use pre-defined rules to recognize patterns but rather learns on its own through training and adjusting parameters), has demonstrated similar performance to attention-based models but with reduced time complexity. This means that it could essentially process longer contexts.
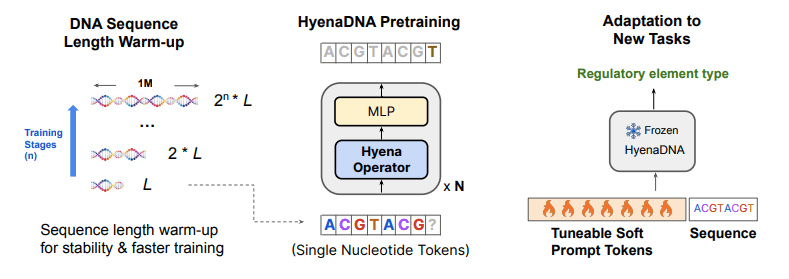
Image Source: https://arxiv.org/abs/2306.15794
Hyena implements a global convolutional filter (a tool that helps a model look at all the parts of the information at once) and a data-controlled gating mechanism (A mechanism that allows the model to decide how much attention to give to which part of the data based on particular characteristics of the data) to perform a context-specific operation on each and every individual token. With these core operations of Hyena, Eric Nguyen and his team developed a genomic foundation model called HyenaDNA. The model was pre-trained on the human reference genome at context lengths that go up to 1 million tokens and a resolution on the level of a single nucleotide (500x increase over attention-based models). HyenaDNA can deal with longer sequences much better because it scales sub-quadratically rather than quadratically. Moreover, it employs single nucleotide tokens and captures information from the entire input at each layer.
The modeling begins with a decoder-only Hyena architecture, which is trained by trying to predict the next nucleotide in a sequence based on the patterns it has learned. Rather than aggregating tokenizers, a single-character tokenizer and a minimal DNA vocabulary of 4 nucleotides (plus special tokens) are utilized. However, the training encounters instability problems as the sequences exceed a length of 2,00,000. To tackle this problem, a sequence length warm-up scheduler is introduced, which causes a gradual increase in sequence length in stages. The sequence length warm-up scheduler was observed to have reduced training time by 40% and improved accuracy by 7.5 points on a species classification task.
Apart from that, downstream adaptation procedures were also designed to benefit from the longer context window, which included the introduction of a soft prompt technique. The soft prompt technique involves the injection of learnable tokens (up to 32,000) into the input sequence itself. The learnable tokens act as hints or prompts during the processing and foster competitive downstream results without any need for updating the pre-trained model.
So far, the features and capabilities of HyenaDNA seem suitable for effectively processing DNA sequences and extracting vital information from them. But does it deliver on its performance promises? Let’s see.
The Powerful Performance of HyenaDNA Revealed!
When it came to evaluating the performance, HyenaDNA showed superb results. A list of its excellent performances is as follows:
- The researchers applied HyenaDNA models to 28 different downstream genomic processes, demonstrating its long-range proficiency and precise resolution.
- In comparison to the Nucleotide Transformer, HyenaDNA could achieve state-of-the-art (SotA) performance on 12 out of 17 datasets on fine-tuned benchmarks while utilizing substantially fewer parameters and pre-training data.
- On the Genomic Benchmarks, it successfully surpassed all eight datasets with a mean improvement of 9 accuracy points and about 20 accuracy points on identifying enhancer function.
- HyenaDNA effectively classified a previously unknown species by increasing context lengths up to 1 million tokens.
- Even a challenging, mammoth chromatin profiling experiment involving 919 distinct classifications related to chromatin profiles was dealt with by HyenaDNA with such efficiency that it outperformed a larger SotA sparse-attention BigBird Transformer.
- HyenaDNA also demonstrated great results in classifying sequences based on their biotypes (biological properties).
Confronting the Limitations of HyenaDNA
Undoubtedly, HyenaDNA shows remarkable performance, but it is currently in its early stages. Some hurdles that still need to be crossed are:
- HyenaDNA has been pre-trained on the reference genome of only a single human. In order to improve the generalizability of features and lessen bias, it must be trained on genomes other than those of humans.
- The current focus of HyenaDNA is on DNA only. The capabilities of HyenaDNA should be extended to other biomolecules such as RNA or proteins as well.
- And lastly, the model size of HyenaDNA is smaller than that of the previous models. Increasing the size could provide additional capabilities.
Even with these shortcomings, there is ample scope for improvement for HyenaDNA in the future.
Conclusion
With the advent of HyenaDNA, the future of sophisticated genomic research and development doesn’t seem distant. Its ability to handle long DNA sequences with single nucleotide resolution opens up new possibilities for unraveling the mysteries of the genome and advancing various fields of biological research. It may have certain shortcomings, but with further advancements, it can offer personalized analysis of whole patient genomes, enable the design of synthetic regulatory elements, genes, and protein complexes, and even unleash the next wave of innovation in AI-based drug discovery and drug design. Ultimately, HyenaDNA has the potential to become a one-stop solution to decoding life itself!
Article Source: Reference Paper | HyenaDNA Availability: Hugging Face | Colab | GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Neegar is a consulting scientific content writing intern at CBIRT. She's a final-year student pursuing a B.Tech in Biotechnology at Odisha University of Technology and Research. Neegar's enthusiasm is sparked by the dynamic and interdisciplinary aspects of bioinformatics. She possesses a remarkable ability to elucidate intricate concepts using accessible language. Consequently, she aspires to amalgamate her proficiency in bioinformatics with her passion for writing, aiming to convey pioneering breakthroughs and innovations in the field of bioinformatics in a comprehensible manner to a wide audience.










