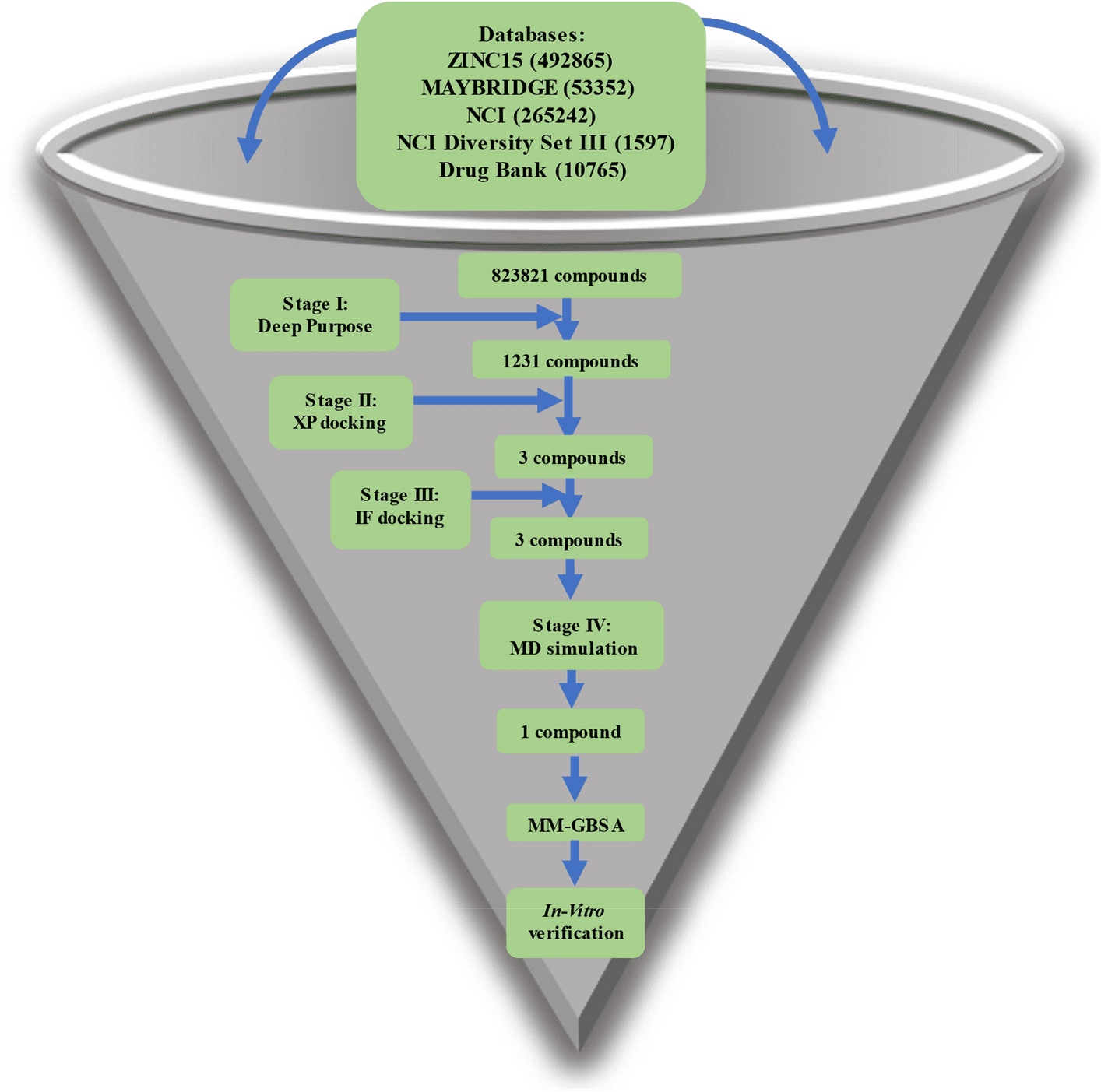
Since the first appearance of SARS-CoV-2, a number of in silico methods have been employed to discover possible drugs that can target the viral life cycle. Scientists from National Research Centre, Egypt, targeted Endoribonuclease, which is one of the crucial non-structural proteins since it is responsible for processing viral RNA to escape detection by the host defense system. The research explores a hierarchical structure-based virtual screening method to target Endoribonuclease or NSP15 of SARS-CoV-2.
Based on the information provided by the International Committee on Virus Taxonomy (ICTV), Coronavirus belongs to the Coronaviridae family and the Riboviria realm. Currently, there are seven strains of the virus that can cause illness in humans, and it was discovered in human beings for the first time in the 1960s. According to the sequence similarity analysis, SARS-CoV-2 shares 79% similarity with SARS-CoV and 50% similarity with MERS-CoV. SARS-CoV-2 shares more similarities with two bat CoV strains than SARS-CoV. There is a 96.2% similarity between RaTG13, which was discovered in Rhinolophusafnis, and SARS-CoV-2, which was discovered in humans in 2009.
Deep learning, which is a branch of machine learning, uses artificial neurons to process data. There have been applications of deep learning (DL) in two sectors: text mining and picture pattern recognition. Additionally, virtual screening, molecular docking, and QSAR models are used to speed up drug discovery processes. Researchers have used deep learning algorithms to identify Coronavirus infections on radiographs, find new antibiotic compounds, and predict drug-target interactions. Prior to the use of deep learning, drugs must be vectorized in order to map their properties to the vectors. With deep learning, large-scale data can be predicted more quickly than with traditional drug discovery methods.
In addition to its (+) sense RNA genome, SARS-CoV-2 is also a beta-coronavirus with a genome of nearly 30 kb. Spike (S), envelope (E), membrane (M), and nucleocapsid (N) are the four structural genes encoded in this genome. Then there are nine accessory genes (ORF3(a, b), ORF6, ORF7(a, b), ORF8, ORF9(b, c), and ORF10) which encode the 16 non-structural proteins (NSPs). The N-terminal domain, the middle domain, and the C-terminal domain make up the endoribonuclease (EndoU or NSP15), a 346 residue protein. RNA replication and transcription are facilitated by the protein’s C-terminal domain, whereas hexamers are formed by its N-terminal domain.
Single-stranded RNA molecules containing uridine (U) sites are hydrolyzed by NSP15 by hydrolyzing their phosphodiester bonds. By cleaving this bond, 2′, 3′ cyclic phosphodiesters are produced along with 5′ hydroxyl terminals. SARS-CoV-2 virus replication would not be possible without the PolyU sequence in the negative-sense viral RNA replicated from polyadenine. Without this structure, the host’s innate immune system would recognize the negative-sense viral RNA using the pattern recognition receptor MDA5. By inhibiting cytoplasmic stress granule formation with antiviral function, this discovery leads to the RNA-activated antiviral response.
Furthermore, deletion of NSP15 is reported to decrease viral replication significantly. The NSP15 protein also inhibits interferon-beta (IFN-β), the production of which is associated with retinoblastoma tumor suppressor protein. The SARS-CoV-2 virus may target this protein due to its vital role in virus replication.
The authors used four different drug databases to target NSP15, an endoribonuclease. Prior to molecular docking, a state-of-the-art Deep Learning library (Deep Purpose) was employed to screen and filter the compounds. The Glide tool under Schrödinger’s equation was used to dock the compounds in two steps via Extra Precision (XP) mode and Induced Fit (IF) mode. Molecular Dynamic Simulations (MD) were carried out in triplicate for 100 ns each. Using Molecular Mechanics-Generalized Born Surface Area (MM-GBSA), binding energy was calculated for each compound after MD simulation and for each amino acid’s contribution to the binding. A series of in vitro tests were undertaken to find the 50% inhibitory concentration (IC50), 50% cytotoxic concentration (CC50), and the selectivity index (SI) of this new compound.
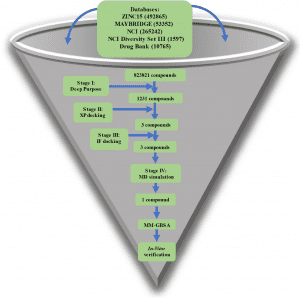
Image Source: https://doi.org/10.1038/s41598-022-17573-6
A directed message passing neural network has been developed by Wang et al. In order to screen drugs against SARS-CoV-2 NSP5. To fine-tune the model, the researchers trained it on experimental data related to several beta CoVs, followed by drugs discovered which were active and inactive against SARS-CoV-2. A total of 5 million drug-like compounds from the ZINC15 dataset were screened using the final model.
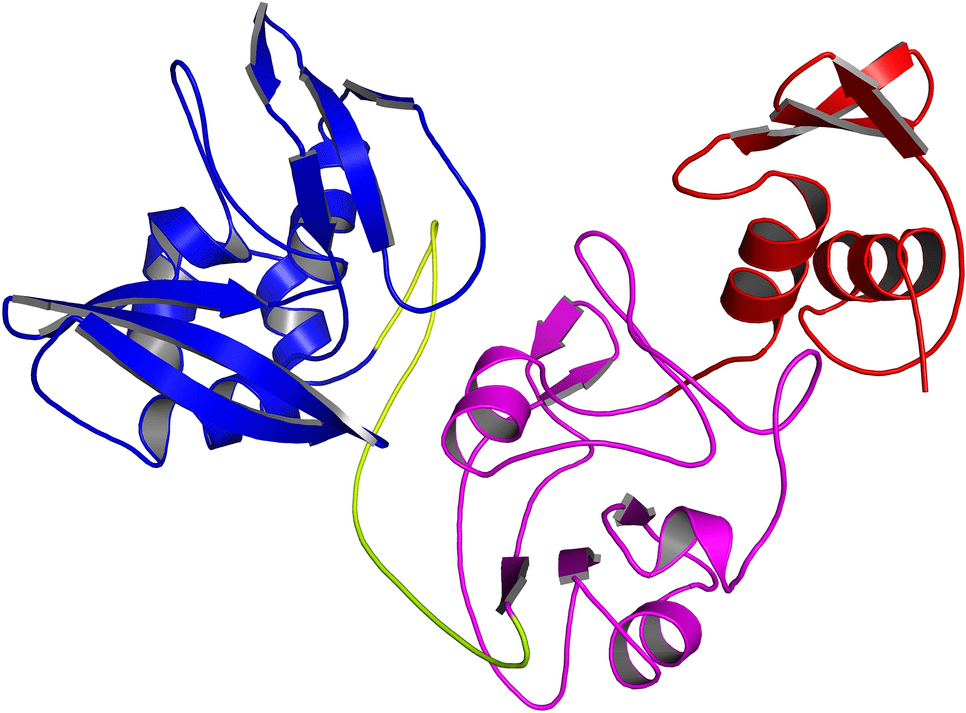
Image Source: https://doi.org/10.1038/s41598-022-17573-6
Various in silico techniques have been used over the past few years to identify potential drugs that can weaken the viral life cycle since SARS-CoV-2 first emerged. These techniques range from classical virtual screening and molecular docking techniques to deep learning models that employ state-of-the-art algorithms.
According to El-Demerdash, A. et al., 15 guanidine alkaloids were screened against two NSPs (NSP10 and NSP5), three structural proteins (S-N-M), and their structure-activity relationships (SAR) were determined, and their properties of absorption, distribution, metabolism, excretion, and toxicity were predicted. As a result of the screening, two compounds showed positive results against their target proteins (crambescidin 786 and crambescidin 826).
In a pharmacophore-based screening, ElHassab et al. found only 24 drug-like compounds from 48 million in the ZINC database. Following the docking scores of the target protein, these compounds were further divided into four groups for MD simulations. According to their results, only one compound displayed good binding affinity in comparison to Sinefungin, a pan-methyl-transferase inhibitor, and might inhibit NSP16.
In SARS-CoV-2, NendoU and NSP15 are the two major RNA uridylate-specific endoribonucleases. There is a specific catalytic site for uridine at its C-terminus. The role of NSP15 in innate immunity has recently been discovered, so it is thought to be a crucial target for fighting infection. An Mn+2-dependent activity specifically cuts double-stranded RNA. There are six key amino acids within the C-terminal domain (H235, H250, K290, S294, T341, and Y343), provided by two antiparallel beta-sheets.
Although we have been vaccinated against SARS-CoV-2, the virus continues to affect our lives on a daily basis. In order to inhibit viral spread, potential therapeutics are urgently needed.
Article Source: Ibrahim, I. M., Elfiky, A. A., Fathy, M. M., Mahmoud, S. H., & ElHefnawi, M. Targeting SARS-CoV-2 endoribonuclease: a structure-based virtual screening supported by in vitro analysis. Scientific Reports, 12(1). (2022). https://doi.org/10.1038/s41598-022-17573-6
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.