Scientists at Westlake University have developed FoldToken2, a novel approach to protein structure representation that is capable of surpassing the coordinates. This innovative method transforms complex 3D protein structures into a series of tokens, like words used in a sentence, to represent key characteristics of protein structure. FoldToken2 works as the Rosetta Stone for proteins, enabling scientists to think and design them from scratch. This may change protein science forever and transform medicine and material science or even serve as a springboard for future scientific breakthroughs into uncharted territories. Get ready to dive into the enthralling world of FoldToken2 and consider how it has revolutionized protein design!
Introduction
Proteins are very important in performing thousands of activities that are necessary for life. However, their secret is hidden in their complex three-dimensional shapes, which determine their interactions with other molecules. To make breakthroughs in medicine, material science, and many more, we need to understand these structures. ESMFold, AlphaFold2, and AlphaFold3 are some of the deep learning-based methods that have proved useful tools with impressive accuracy in predicting protein structures. These predictions are often guided by multiple sequence alignment (MSA) as well as available protein structure databases. Nevertheless, even such advanced techniques fall short when dealing with 3D coordinates’ inherent equivalence. Despite this lack of alteration in overall shape, slight rotations or translations in a protein structure can cause significant changes to its coordinate data. For example, it becomes difficult to use these techniques on tasks like protein structure comparison or analysis. This is where FoldToken2, an improved version of FoldToken1, comes in, which creates a unique ‘language’ that can represent protein structure. This new method could potentially change and enhance protein science greatly.
The Challenge: Capturing the Essence in Numbers
Think of proteins as an origami sculpture. The folds and bends in proteins determine their purpose. Traditionally, scientists have represented such structures using 3D coordinates of each atom within the given protein molecule. Although this method provides accurate details, it becomes difficult to use when comparing different proteins or designing new ones.
The flaw lies with “equivalence” in 3D coordinates. In other words, imagine rotating your origami sculpture – all individual coordinates will be changed while the general structure remains the same. Existing methods do not efficiently recognize these interchanges, making it hard to develop algorithms that can quickly analyze and manipulate protein structures.
FoldToken2: A New Way to Speak Protein
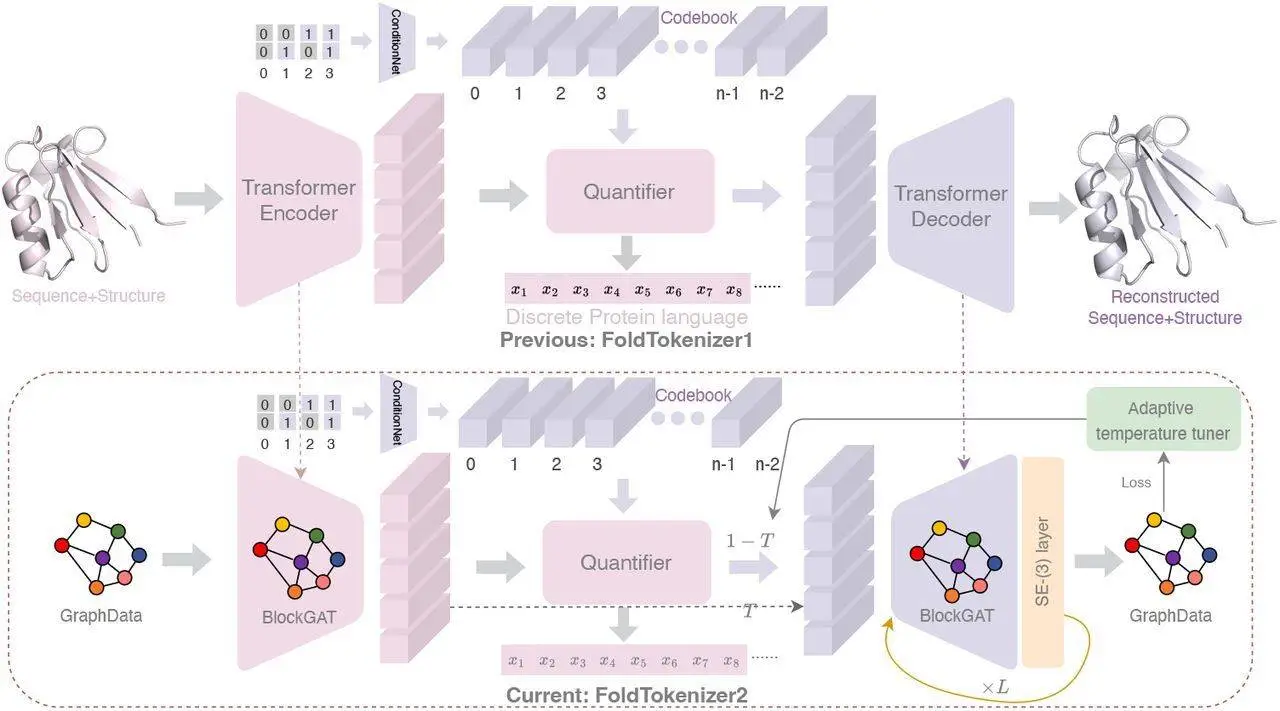
FoldToken2 aims to address this by proposing a fresh language designed explicitly for protein structures. Instead of using raw coordinates, the sentences in this language make use of discrete tokens that are equivalent to words in sentences.
The following is how it happens:
Encoding the Structure: FoldToken2 aims to address the issue of equivariance by focusing on capturing the inherent properties of a protein’s structure that remain unchanged despite its orientation in 3D space. This would enable the system to create a more compact and coherent model for analyzing and manipulating proteins. BlockGAT, a specific kind of neural network layer, is used to analyze block graphs and learn higher-level representations of protein structure. The layer considers the local features of each amino acid block, their relative positions, and transformations (rotations and translations).
Quantization: Turning Numbers into Tokens – Next, the encoded information is processed with the help of a technique called “SoftCVQ.” Consider a large vocabulary of words – SoftCVQ selects one that best represents an encoded protein structure from this list (word). Consequently, this codification gives way to a compressed token sequence representing the whole protein structure.
Decoding: Back to Structure – The last stage requires the conversion of these tokens back into a 3D structure to produce an efficient plug-and-play SE(3)-layer that could be added to any GNN layer for structure prediction. Thanks to the simplified module of the SE(3)-layer and BlockGAT with sparse graph attention, researchers were able to train the model on the entire PDB dataset in 1 day using 8 NVIDIA-A100s. This implies that FoldToken2 has a special decoder that gradually improves its initial guess so that it becomes similar again to the given protein.
The Power of FoldToken2
FoldToken2 offers several advantages over traditional methods:
Compactness: Tokens allow the proteins to be represented in a manner that is more efficient than storing extensive 3D coordinates. It enables quick manipulation and analysis of vast amounts of data.
Invariance: The language of FoldToken2 for protein structures is invariant under small rotation or translation. This implies that even if the raw coordinates change slightly, the sequence of tokens remains unaltered, thereby capturing the true form of the protein.
Generativity: Besides representing known protein structures, the system can generate new ones according to its learned token language. This opens possibilities for creating novel proteins tailored for medical use or materials science.
FoldToken2: The Future of Protein Science in a Nutshell
Protein science is about to undergo a major revolution if this innovative approach is anything to go by. FoldToken2, a compact invariant generative language that deals with protein structures, could speed up the analysis of protein structure, develop new protein-based drugs, or even open doors for designing materials with special functions.
Protein structure analysis can be accelerated by FoldToken2, which provides a concise, invariable, and productive language for them; it can help to establish new drugs that are based on proteins and pave the way for designing materials with specific functions. Interestingly enough, this language allows the creation of entirely new protein structures. FoldToken2 has shown considerable improvement in the quality of structure prediction as compared to other methods, such as TMScore and RMSD, which increased by 20% and 81%, respectively. Moreover, FoldToken2 has also implemented this idea for multiple polypeptide chains’ structure prediction and produced highly reliable results. Such advances could facilitate breakthroughs in areas like enzyme development and even create new biomaterials. The introduction of FoldToken2 will provide more insights into learning protein structural descriptions, alignment, and generation.
Join the Conversation: Constructing the Future of Protein Design
FoldToken2 represents an important development in our understanding and manipulation of proteins. We can expect better refinement and integration with other protein modeling techniques with time. This means that more options are open to us regarding what may happen next.
What are your thoughts on FoldToken2 and its potential impact? Let’s hear what you have to say; comment below!
Article Source: Reference Paper | FoldToken2 is available on Zenodo | Online example is available at Colab.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.