Shine is a computational approach for extracting distinct, well-conserved biomarkers from a large dataset of the microbial genome. The method has been clinically tested and showed promising results in detecting all pathogenic microorganisms with lower false positives or false negatives rates. Shine utilizes a two-directional approach to identify consensus sequences by identifying distinct regions or sensitive regions to successfully determine all pathogenic microorganisms.
Need for early detection of pathogen
Microbial infections are the leading cause of morbidity around the world. With around 1400 known disease-causing pathogens, a lack of rapid and reliable detection methods poses a great problem, as it is the single most important step of diagnosis. The genomic sequence plays a major role in the identification of microbes, and polymerase chain reaction (PCR) is the tool of choice for nucleic acid-based diagnostics. Ribosomal DNA (rDNA) also plays an important part in inferring the reliable identification of pathogens. The pressing need to develop fast detection and reliable pathogen screening methods in a cost-effective way has led to the further improvement of electrochemical detectors for microorganisms. DNA samples obtained from disease-causing microorganisms are in very low quantity. Traditional PCR and real-time PCR over time have been found to have low pathogen detection sensitivity. Other methods pose a high risk of contamination, like a two-step nested polymerase chain reaction (PCR). Apart from it, these methods are often costly and time-consuming.
Biomarkers from plasmids and 16S Ribosomal RNA (16S rRNA)
To identify biomarkers as templates in order to design probes and primers for identifying microbes, particular sequences obtained from plasmids and 16S ribosomal RNA (16S rRNA) are used as targets. Biomarkers obtained from plasmids and the 16S rRNA of microorganisms still need laboratory validation. Since rRNA exists in the genomes of all microorganisms, 16S RNA gene sequence analysis is a commonly used method to detect pathogens in samples with a higher sensitivity rate, as they are usually present in multiple copies. However, all rRNA genes may not be species-specific, and it may be challenging to recognize the conserved regions. On the other hand, not all microorganisms have plasmids, and those that contain plasmids may not be species-specific. Hence, neither the 16S RNA gene sequence nor plasmids can be used to design probes and primers to detect pathogenic microbes effectively. The pathogenic genomic data included in Shine was derived from public databases that contained incompletely assembled motifs along with completely assembled disease-causing microbe genomes.
Shine technology
Scientists have developed a de novo genome sequencing-based pipeline to identify original as well as distinct multicopy biomarkers in a microorganism population. The proposed Shine strategy is a comparative genomics-based pipeline of genome alignment that is helpful in analyzing both original as well as distinct biomarkers for an organism. The effectiveness of biomarker detection is directly related to the availability of genomic data. Shine strategy could identify obscure fragments to act as target genes from species-specific microorganisms to form templates and further aid the production of probes and primer sets. The Shine strategy is based on the blast (Basic Local Alignment Search Tool) algorithm for the segregation process to explore the original as well as particular repetitive biomarkers in the microbe population. This technology has been shown to display low rates of false positives or negatives and is applicable for PCR primers and various other probes. The Shine technology has evolved from molecular data sequencing tools like Phylogenetic Analysis by Maximum Likelihood (PAML4) and Molecular Evolutionary Genetics Analysis (MEGA5). Shine is imperative for carrying out comparative genomics studies in routine tests for better efficiency in the early detection of microorganisms.
Image source:https://doi.org/10.1186/s12859-023-05195-2
Why use Shine?
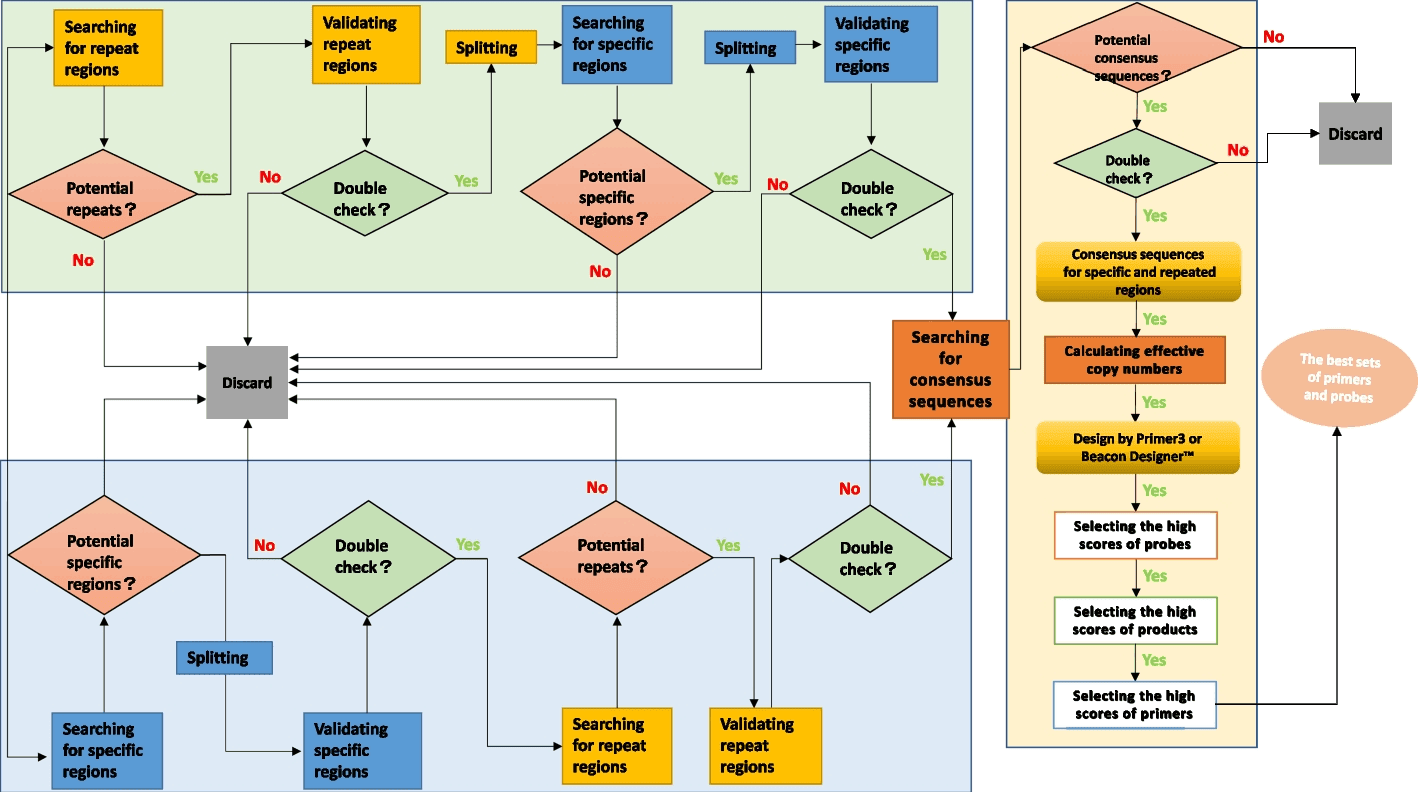
Shine pipeline is a two-pronged approach wherein the available sample data sequence, whether it is specific regions or repetitive regions, was divided into two selective procedures. For organisms with smaller genomes, specific gene sequence was searched, whereas for large genomes, multicopy regions were searched for. Later, the data from the searches were used to find consensus regions in the sequence to be able to design the perfect probe and primer set. Subsequently, double-check verifications were carried out in every module to check the efficacy of the method.
The above illustration shows a subpart of the workings of search methods to sort specific regions from sample data. The target sequence obtained from microorganisms is initially compared against the complete sequence of the whole genome of one or more strains available in the database. This is done to obtain cut fragments in the first round. This is followed by comparing the target sequence with the rest of the remaining strains in order to obtain organism-specific fragments, followed by validation of specific regions as possible biomarkers. Apart from it, the copy number of different repetitive regions from the target sequence is identified along with alterations in repeats so as to discard the rest of the noise data. The sequences thus obtained undergo validation, where they are remapped to verify the accuracy of the data. Potential repeats are statistically analyzed to find the target sequence’s mean copy number of repetitive regions.
Often, target sequences obtained from microbes have various incomplete motifs. So, while identifying distinct multicopy biomarkers of bacterial origin, the motifs of target microbial sequences are first connected together. The Shine pipeline initially aligns the target sequence internally to identify multicopy regions.
Conclusion
The Shine pipeline is remarkably sensitive and may be capable of detecting newly discovered multicopy biomarkers specific to species, even subspecies-specific target sequences, including all identified epidemic-causing microorganisms. The Shine strategy is best suited to identify shared universal phylogenetic biomarkers with fewer false negative or positive results and help in designing primers and probes to detect disease-causing microorganisms. Once the biomarkers have been identified, graphene arrays could be used to detect various strains, e.g., the application of graphene-based arrays has been used for detecting viruses like SARS-CoV-2, dengue virus, influenza, hepatitis C, etc.
Article Source: Reference Paper | SHINE: Website
Learn More:
Sipra Das is a consulting scientific content writing intern at CBIRT who specializes in the field of Proteomics-related content writing. With a passion for scientific writing, she has accumulated 8 years of experience in this domain. She holds a Master's degree in Bioinformatics and has completed an internship at the esteemed NIMHANS in Bangalore. She brings a unique combination of scientific expertise and writing prowess to her work, delivering high-quality content that is both informative and engaging.