Chromatin conformation capture techniques have revolutionized our understanding of genome organization and its impact on gene regulation. Hi-C, the gold standard method for investigating chromatin contacts, has shed light on the three-dimensional organization of the genome. Nonetheless, new developments in genomic technology have opened the door for new strategies that offer a more thorough understanding of chromatin interactions. One such breakthrough is the Multiplex-GAM (genome architecture mapping) method, which goes beyond the limitations of Hi-C and unveils new insights into the intricacies of chromatin contacts.
Understanding Hi-C and Its Limitations
Hi-C is a powerful technique that investigates genome-wide chromatin contacts by cross-linking and sequencing DNA fragments that are in close proximity to each other. By analyzing the frequency of interactions between different genomic regions, Hi-C enables the construction of three-dimensional maps of chromatin interactions.
However, Hi-C has some limitations, such as it is a lengthy and expensive process that requires a large amount of input DNA. Additionally, Hi-C suffers from limited resolution, as the generated maps are dependent on the size of the DNA fragments sequenced. This limits the detection of interactions occurring at smaller scales, such as within gene regulatory elements and enhancer-promoter loops. Furthermore, Hi-C predominantly captures long-range interactions, which may overshadow the significance of short-range interactions within the same genomic region.
Introducing Multiplex-GAM
Multiplex-GAM is an innovative approach that overcomes the limitations of Hi-C and allows for a more comprehensive exploration of chromatin contacts. This method utilizes proximity ligation to capture DNA fragments that interact in close proximity, followed by amplification and high-throughput sequencing.
One key advantage of Multiplex-GAM is its ability to detect short-range chromatin contacts with higher resolution. By utilizing shorter DNA fragments, Multiplex-GAM enables the identification of interactions occurring within gene regulatory elements and enhancer-promoter loops that may have been missed by Hi-C. The spatial arrangement of the genome and how it affects gene expression can be better understood with increased precision.
Another notable feature of Multiplex-GAM is its capacity to capture multi-way chromatin interactions. While Hi-C primarily captures pairwise interactions, Multiplex-GAM can identify simultaneous interactions between multiple DNA fragments. This allows for the discovery of complex chromatin architectures, such as chromatin hubs or regulatory networks involving multiple genes.
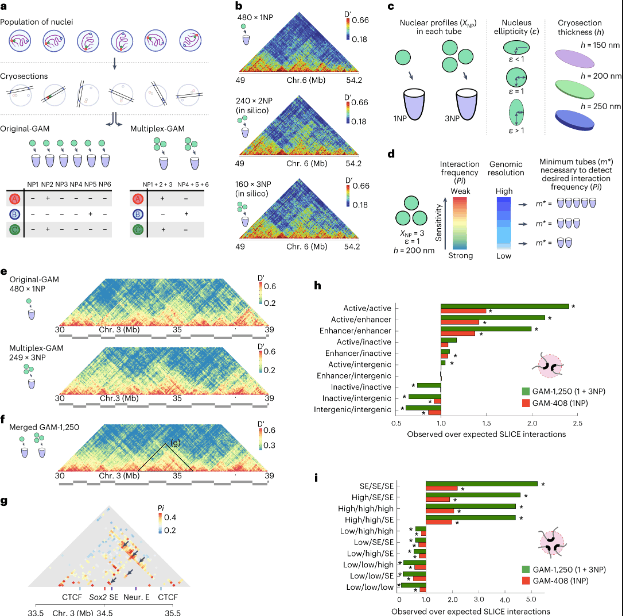
Image Source: https://doi.org/10.1038/s41592-023-01903-1
Complex Insights Uncovered by Multiplex-GAM
Multiplex-GAM has already provided valuable insights into chromatin contacts that Hi-C previously overlooked. In genome-wide 3D chromatin folding assays, the detection of topologically associating domains (TADs) is a crucial objective. A comparison was conducted between TAD boundary calls using GAM, bulk Hi-C, and single-cell Hi-C. The results revealed that all three approaches identified similar boundaries, with those detected by all methods exhibiting stronger insulation. It was noted that unique TADs identified by specific methods likely represent method-dependent variability. Furthermore, the distribution of previously reported features enriched at TAD boundaries was generally similar between Hi-C and GAM, although a few epigenetic features were not found to be enriched in the small subset of boundaries unique to GAM.
Furthermore, Multiplex-GAM has shed light on the dynamics of chromatin contacts during development and disease. By comparing chromatin interaction maps across different cell types or disease states, researchers have discovered specific alterations in chromatin contacts that contribute to the regulation of gene expression.
Additionally, Multiplex-GAM has unveiled novel insights into enhancer-promoter interactions. By analyzing the fine-scale contacts within gene regulatory elements, researchers have gained a deeper understanding of how enhancers and promoters physically interact to regulate gene expression. These discoveries have significant implications for understanding cellular identity, differentiation, and disease pathways.
Researchers applied multiplex-GAM to various neuronal subtypes in brain tissues, revealing unexpectedly extensive chromatin decondensation in long neuronal genes. They also identified cell-type specific contacts that encompass differentially expressed genes and accessible regulatory elements spanning multiple megabases.
Conclusion
Multiplex-GAM represents a significant advancement in the field of chromatin conformation capture. By surpassing the limitations of Hi-C, Multiplex-GAM allows for a more comprehensive exploration of chromatin contacts, unveiling hidden insights into genome organization and gene regulation. The ability to detect short-range interactions with higher resolution, capture multi-way interactions, and study non-coding regions and allele-specific interactions opens up new avenues for interpreting the spatial organization of the genome and its functional consequences.
The insights gained from Multiplex-GAM have the potential to drive breakthroughs in fields such as developmental biology, disease research, and personalized medicine. As genomic technologies continue to evolve, it is exciting to envision the discoveries that will emerge from this innovative method and its applications in unraveling the mysteries of chromatin contacts.
Article Source: Reference Paper
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.