Cell-cell interactions (CCIs) are pivotal in various biological functions, including cellular differentiation, tissue maintenance, and immune reactions. As single-cell RNA sequencing (scRNA-seq) techniques advance, the need to discern CCIs from the growing pool of data becomes crucial. A groundbreaking technique, “DeepCCI,” developed by researchers from the Harbin Institute of Technology, China, uses scRNA-seq data to explore intercellular communication in higher multicellular organisms, i.e., animals. DeepCCI showcases its remarkable versatility by successfully applying its capabilities to a diverse array of publicly available datasets across various technologies and platforms. This demonstrates its exceptional accuracy and effectiveness in predicting significant CCIs. With its user-friendly software, DeepCCI emerges as a comprehensive solution, streamlining the process of uncovering meaningful intercellular communications and constructing CCI networks from scRNA-seq data.
Single-cell RNA sequencing innovations have made significant commitments to figure out the cellular mechanisms. They permit the identification of distinct cell types and states, help in supporting personalized medication procedures biomarker revelation, upgrade insights into advancement, and extend our cognizance of illness systems. In spite of the difficulties in examining scRNA-seq information, the potential for extraordinary bits of knowledge into cell components and the treatment of different illnesses is significant.
Importance of DeepCCI
The application of DeepCCI to various scRNA-seq datasets demonstrates its effectiveness in efficiently clustering different cell types and forecasting their communication patterns. With the aid of this technology, it is now possible to efficiently comprehend basic biological processes and decipher the intricate operations of cells in complex animals. The researchers also made a comprehensive ligand-receptor pair database called the Ligand-Receptor Interaction Database (LRIDB).
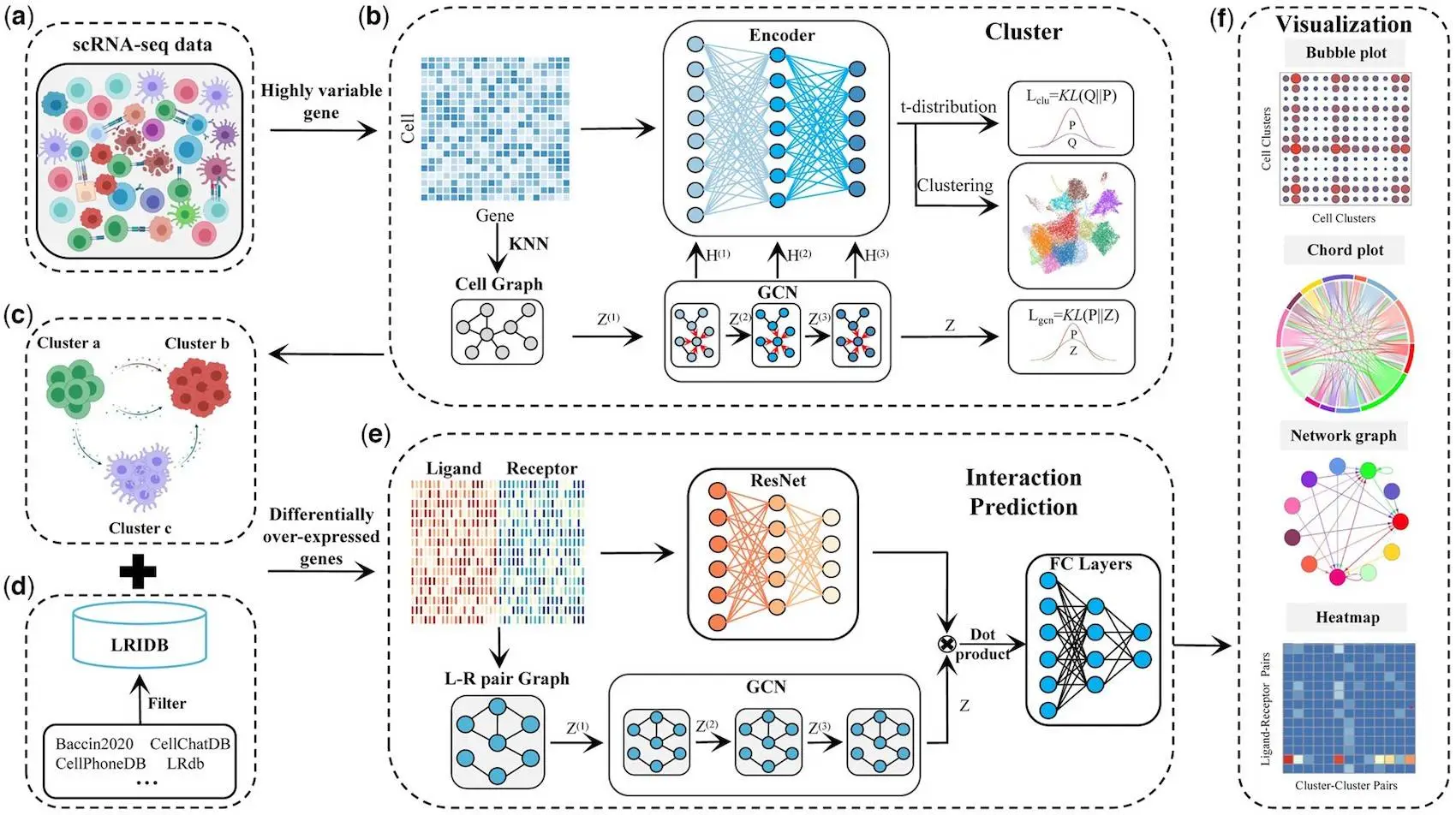
To explore the interactions, DeepCCI provides two deep learning models: (i) a GCN-based unsupervised model for cell clustering and (ii) a GCN-based supervised model for CCI identification. DeepCCI has great potential to cluster cells and capture meaningful biological interactions between cell clusters by utilizing heterogeneous cells’ underlying complex gene expression patterns from scRNA-Seq data. DeepCCI provides several visualization outputs to show how strongly or specifically each cell cluster interacts with every other cell cluster.
Workflow of the Model
The workflow of DeepCCI involves constructing an extensive ligand-receptor pair database (LRIDB) and preprocessing the scRNA-seq dataset to ensure relevance and quality. While many databases exist to provide this information, researchers built one, called LRIDB, by carefully choosing and curating L-R pairs from various sources to ensure broad coverage and biological relevance.
The data from the scRNA-seq experiment was then ready for analysis. Only relevant genes expressed in at least 1% of cells and cells expressed in at least 1% of genes were kept after data quality was filtered and controlled. The top 2000 genes were chosen for the study after the data had been normalized and based on variance.
The technique was built around two key components: autoencoders (AE) and graph convolutional networks (GCN). The AE aided in the learning of single-cell expression data representations, responding to different data properties. It was made up of an encoder and a decoder that worked together to recreate input data properly. The GCN, on the other hand, made use of cellular relationship topology. It propagated information via the cell and L-R pair graphs to update node representations. This enabled the model to understand the complicated connections between cells, which aided in the prediction of significant interactions.
DeepCCI combines several approaches to cluster cells and anticipates intercellular connections, yielding significant insights into complicated biological processes that rely on coordinated cellular activity. Hence, it proved DeepCCI’s effectiveness on scRNA-seq datasets.
Model construction for single-cell clusters
The scientists used a graph-based methodology to categorize individual cells based on their gene expression patterns. It started with a cell network in which cells were nodes and edges represent interactions between cells determined by their K (default value of 10) nearest neighbors. Then, the Autoencoder (AE) and graph convolutional network (GCN) were merged to generate a complete cell representation for enhanced grouping. The AE is pre-trained to reconstruct the gene expression matrix, aiding subsequent clustering. The interaction networks are established by identifying differentially overexpressed L-R pairs in cell clusters and predicting significant CCIs using a Residual Network (ResNet) and GCN model. Pretraining the model improved performance substantially. A probability distribution was created to allocate cells to clusters and optimize the data representation to match cluster centers. These assignments were used to generate the final clustering findings, allowing for effective cell grouping.
Identification of significant interactions
To determine significant interactions between cell clusters mediated by ligand-receptor pairs, the researchers calculated interaction probability values based on the average expression levels of ligands and receptors in the respective cell groups. Further, statistical methods like CellChat, CellPhoneDB, and SingleCellSignalR were used by applying a predefined threshold for significance. The significant interactions identified by majority vote were used as the true labels for the model.
Building the DeepCCI Model and Establishing Evaluation Benchmarks
DeepCCI, a predictive model for cell-cell interactions, combines a Residual Network (ResNet) and Graph Convolutional Network (GCN). ResNet takes ligand and receptor expression values from cell clusters as input, utilizing geometric mean for multi-subunit proteins. Singular Value Decomposition (SVD) is used to reduce feature data dimensionality. GCN processes the ligand-receptor (L-R) pair graph based on scRNA-seq data and LRIDB. The output of ResNet and GCN is combined and fed to fully connected layers for predicting L-R interactions. Focal loss is employed for network training. Evaluation metrics include Adjusted Rand Index, recall, precision, accuracy, F1-score, and Area Under the Curve (AUC) for performance assessment.
Model Evaluation
Method comparisons of the DeepCCI cluster model
To assess DeepCCI’s cell clustering performance, it was compared with 13 state-of-the-art methods on 12 real-world scRNA-seq datasets using the Adjusted Rand Index (ARI), a common clustering metric. The comparison involved averaging results from 10 runs of each method to ensure robustness and accuracy in the evaluation.
Assessment of DeepCCI’s Interaction Model
The evaluation of DeepCCI’s interaction model involved multiple datasets. The primary dataset, panc8, integrated eight pancreas datasets across five sequencing technologies. Additionally, two independent test sets were used: a human atopic dermatitis (AD) dataset with 17,349 cells clustered into 12 groups and an embryonic mouse skin dataset with 25,148 cells clustered into 13 groups. For further validation, three diverse datasets were used: a scRNA-seq dataset of human testicular cells, a seqFISH dataset of mouse organogenesis, and a 10x Visium spatial transcriptomics dataset of the mouse brain. To assess spatial influence on interactions, neighboring cell types were considered. Lastly, DeepCCI was applied to the PBMC3k scRNA-seq dataset to demonstrate its capability for de novo prediction of cell clustering and interactions. Various evaluation metrics were utilized, including AUC, precision, and ARI, depending on the dataset and context.
Performance evaluation of the cluster model of DeepCCI
The performance evaluation of the cluster model of DeepCCI involved comparing its cell clustering results with various scRNA-Seq analytical frameworks using different metrics. Across 12 scRNA-seq datasets, DeepCCI consistently achieved the best Adjusted Rand Index (ARI) and demonstrated stability in cell clustering as shown by other metrics like Normalized Mutual Information (NMI), Silhouette score, and AMI. Visualization of cell clustering results through UMAPs highlighted the accuracy and ability of DeepCCI to capture hidden cellular information and accurately predict cell clusters. The choice of the number of clusters was identified as a critical parameter affecting clustering performance.
Performance Evaluation of Cluster and Interaction Models in DeepCCI
DeepCCI performed exceptionally well in both cell clustering and cell-cell interaction (CCI) prediction. Its strong clustering outperforms previous approaches, as evidenced by high Adjusted Rand Index (ARI) values and automated cluster number selection. L-R interactions are highlighted using visualization approaches such as bubble plots and network graphs. DeepCCI effectively discovered physiologically meaningful interactions for CCI prediction, which was confirmed using rigorous 5-fold cross-validation and key measures such as F1, Recall, and AUC. Overall, DeepCCI demonstrates proficiency in analyzing complicated intercellular connections, which is critical for comprehending cell interactions in biological systems.
DeepCCI, a state-of-the-art tool, enlightens intercellular correspondence in multicellular organic entities utilizing scRNA-seq information. It utilizes profound learning, especially chart convolutional networks, to bunch cells in light of quality articulation designs. Thus, it predicts connections between cell bunches utilizing an organized sub-atomic collaboration data set. The use of DeepCCI across different scRNA-seq datasets exhibits its ability to group cell types and anticipate their correspondence designs successfully. This tool is essential for disentangling the mind-boggling activities of cells in complex organic entities and figuring out major natural cycles.
De novo prediction of CCIs using DeepCCI
DeepCCI is a powerful tool designed to decode cellular communications within single-cell RNA sequencing (scRNA-seq) data. It tackles the challenge of deciphering interactions between cells in the complex world of genomics data. DeepCCI achieves this by first clustering cells into meaningful groups using a sophisticated clustering model. The accuracy of these clusters is measured, and the resulting clustering is represented visually using a technique called UMAP, showing how well the model matched annotated labels.
Conclusion
DeepCCI excels in predicting critical cell-cell interactions with deep learning, which has been tested against measured labels and statistical approaches. Chord diagrams, for example, assist in analyzing these relationships. Its innovation lies in leveraging a deep learning framework for accurate clustering and rapid identification of meaningful interactions from single-cell data. Future aims include integrating spatial transcriptomics, advancing multi-omics integration, and creating user-friendly software for wider applications.
Article source: Reference Paper
Learn More:
Prachi is an enthusiastic M.Tech Biotechnology student with a strong passion for merging technology and biology. This journey has propelled her into the captivating realm of Bioinformatics. She aspires to integrate her engineering prowess with a profound interest in biotechnology, aiming to connect academic and real-world knowledge in the field of Bioinformatics.














[…] detection of plasmids and viruses within sequencing data is critical in genomics as it helps to learn about the complex variety of these mobile genetic […]