The requirement and utility of conventional force field-based folding simulations have been called into question by the overwhelming success of deep learning techniques in protein structure prediction. Researchers from the University of Michigan and the National University of Singapore presented a hybrid method called deep-learning-based iterative threading assembly refinement (D-I-TASSER), which combines iterative threading fragment assembly simulations with multi-source deep learning potentials to create atomic-level protein structural models. To automate the modeling of large multidomain protein structures, D-I-TASSER presents a domain splitting and assembly technique. For both single-domain and multidomain proteins, D-I-TASSER performs better than AlphaFold2 and AlphaFold3, according to benchmark tests and the most recent critical evaluation of protein structure prediction, which included 15 trials. Additionally, extensive folding tests demonstrate that D-I-TASSER could fold 81% of protein domains and 73% of full-chain sequences in the human proteome, yielding outcomes that are very similar to those of the previously published AlphaFold2 models. These findings open up a new way to combine deep learning and traditional physics-based folding simulations to produce highly accurate predictions of protein structure and function that can be applied throughout the entire genome.
Introduction
The critical assessment of protein structure prediction (CASP) tests conducted across the community has significantly advanced the field of protein structure prediction. Local structure features, including hydrogen bonds, contact and distance maps, and torsion/dihedral angles, have all been predicted using deep learning. By optimally meeting geometry predictions, usually by nuclear magnetic resonance, full-atom relaxation, quasi-Newton minimization, and crystallography, full-length 3D models are created. AlphaFold2 is an end-to-end learning protocol designed to enhance modeling techniques based on two-stage restraints. Diffusion samples can be used to improve end-to-end learning’s efficacy and generality, according to recent research by AlphaFold3. These deep learning techniques outperformed conventional structural folding techniques based on large-scale physical force field-based simulations, like I-TASSER, Rosetta, and QUARK, in terms of accuracy.
The precise and effective modeling of multidomain proteins, which have numerous domains and use domain-domain interactions to carry out higher-level functions, is a problem for the science of proteins. Although domain-level structures are the focus of advanced techniques in the area, most approaches do not have a multidomain processing module, which makes it challenging to precisely and effectively investigate these complex proteins.
D-I-TASSER Hybrid Pipeline
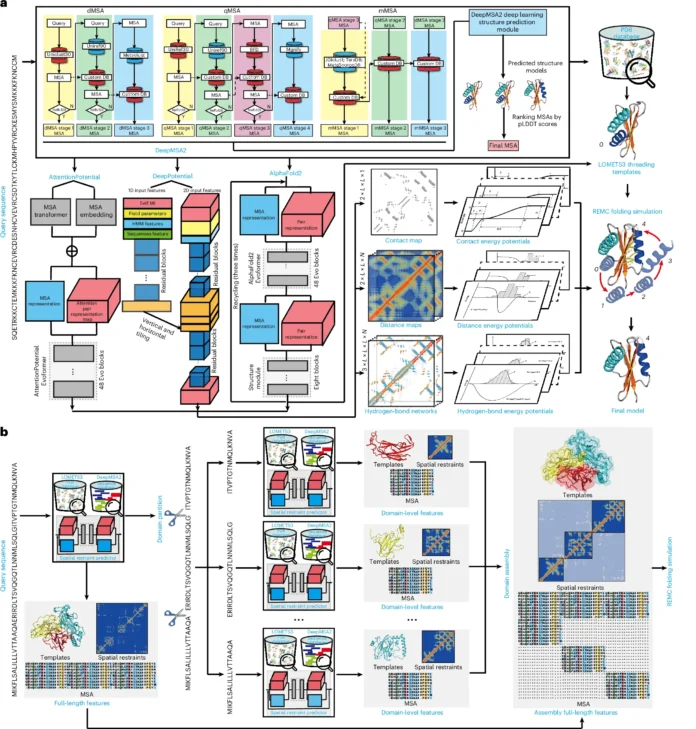
The deep-learning-based iterative threading assembly refinement pipeline (D-I-TASSER) is a hybrid pipeline that combines state-of-the-art iterative threading assembly simulations for atomic-level protein tertiary structure modeling with multisource deep learning features, such as contact/distance maps and hydrogen-bonding networks. Monte Carlo simulations carried out by D-I-TASSER enable the implementation of the full version physics-based force field of I-TASSER for structural optimization and refinement when combined with the deep learning models, in contrast to the quasi-Newton minimization algorithm, which necessitates the differentiability of the objective function. Moreover, a novel domain-splitting and reassembly module is presented for the automated modeling of complex protein structures with many domains.
The hybrid D-I-TASSER pipeline beats the state-of-the-art deep learning techniques AlphaFold2 and AlphaFold3 and outperforms conventional I-TASSER series methods, according to benchmark tests and the most recent blind CASP15 experiment.
In contrast to the newly published AlphaFold Structure Database, D-I-TASSER produced a greater coverage of foldable sequences when used for the structural modeling of the complete human proteome, serving as an example of a large-scale application. The community can now freely access the D-I-TASSER programs and the genome-wide modeling results at https://zhanggroup.org/D-I-TASSER/.
For scholarly purposes, the standalone software and all benchmark datasets are accessible at https://zhanggroup.org/D-I-TASSER/download/.
D-I-TASSER’s Innovative Approach to Domain-Splitting and Assembly for Structural Accuracy
D-I-TASSER is a method for modeling protein structures utilizing hybrid deep learning and threading fragment assembly, with an emphasis on nonhomologous and multidomain proteins. With the help of deep residual convolutional, self-attention transformers, and end-to-end neural networks, it builds deep multiple sequence alignments (MSAs) through quick prediction processes guided by deep learning. DeepPotential, AttentionPotential, and AlphaFold2 are used to create spatial structural restraints. Template fragments from several threading alignments are assembled using replica-exchange Monte Carlo simulations and the LOcal MEta-Threading Server (LOMETS3) to create full-length models. D-I-TASSER includes a new domain partition and assembly module to address the difficulty of multidomain structural modeling. This module iteratively generates threading alignments, domain border splits, domain-level MSAs, and spatial restraints.
First, two sizable benchmark datasets were used to test the pipeline. With an average TM score that is 108% higher than that of the traditional I-TASSER pipeline, D-I-TASSER produces high-quality models for the dataset of 500 single-domain proteins without homologous templates in the PDB, demonstrating a notable influence of deep learning potentials on the folding of nonhomologous structures. With a P value = 1.59 × 10−31 in a paired one-sided Student’s t-test, D-I-TASSER generates full-chain models on the second dataset of 230 multidomain proteins with an average TM score 12.9% higher than that of AlphaFold2 (V2.3), one of the top deep learning techniques in the field.
The new domain-splitting and reassembly protocol, which enables more thorough domain-level evolutionary information derivation and balanced intradomain and interdomain deep learning model developments, as well as more accurate multidomain structural assembly, was shown to have a significant advantage in detailed data analyses.
In the most recent community-wide CASP15 experiment, the pipeline was also tested (as “UM-TBM”). On FM domains and multidomain proteins, respectively, D-I-TASSER achieved the highest modeling accuracy in both single-domain and multidomain structure prediction categories, with average TM scores 18.6% and 29.2% higher than the public March-2022 v.2.2.0 of the AlphaFold2 server run by the Elofsson Lab (registered as “NBIS-AF2-standard”). These findings support the efficacy and promise of physics-based structural assembly simulations for producing high-quality protein tertiary structure predictions when combined with cutting-edge deep learning methodologies.
73% of full-chain sequences (or 81% of domains) can be folded using D-I-TASSER, according to a large-scale practical application that used the program to generate structure predictions for all 19,512 sequences of the human proteome. This information is highly complementary to the recently published human protein models created by the AlphaFold2 program. For structure-based annotation of the multifaceted roles of the proteins in the human genome, these models are thought to be extremely pertinent.
Limitations
Even with the achievements, there are still a lot of obstacles in the field. For instance, shallow MSAs continue to exist for certain proteins even after DeepMSA2 was integrated with large metagenomics databases. This is particularly true for proteins from viral genomics, where the rapid evolution of viruses and their broad taxonomic distribution lead to a dearth of homologous sequences in comparison to other taxonomic groups. Furthermore, the challenge of predicting the structure of protein-protein complexes—a serious issue for which there is currently no workable solution—is not covered in this paper.
Conclusion
The researchers created the D-I-TASSER hybrid pipeline, which combines iterative threading assembly simulations with deep learning capabilities to automatically simulate complex multidomain protein structures. Comparing the provided pipeline to the state-of-the-art techniques, it showed advantages in modeling multidomain proteins and difficult targets. Based on the combination of state-of-the-art physics-based folding simulations and sophisticated deep-learning approaches, these achievements point to a promising future for expanding the current protocol to solve the ongoing difficulties in predicting the structures of protein complexes and orphan proteins.
Article Source: Reference Paper | D-I-TASSER is freely available at the website.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.















