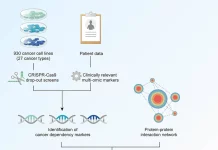
CRISPR-HAWK is a guide-RNA design tool created by researchers from the University of Verona, Harvard Medical School, and the Broad Institute to account for real-world human genetic variation. Unlike typical CRISPR tools, which rely solely on a reference genome, it takes into account population and individual variants, as well as haplotypes, to predict how editing efficiency varies between individuals. Analyzing 79,648 genomes, the scientists discovered that variations can significantly impact guide performance, even preventing cuts at crucial targets like BCL11A, emphasizing the importance of variant-aware CRISPR design.
Overview
CRISPR-Cas systems enable precise DNA editing by employing a guide RNA (gRNA) to drive the Cas enzyme to a target DNA sequence near a PAM (Protospacer adjacent motif) site, and successful editing is dependent on attaining high on-target efficiency while avoiding off-target effects (On-target means editing exactly the right gene. Off-target means accidental editing somewhere else in the DNA). Several computational tools exist to design guide RNAs, including Cas-Designer and CHOPCHOP, which align PAM sites to the reference genome, CRISPick and CRISPRon, which use machine-learning models to rank guides based on efficiency and specificity, and CRISPOR, which annotates guides that overlap known SNPs.
However, because all of these techniques rely heavily on the human reference genome, they simply neglect genuine genetic variation among individuals and populations. For example, Cas-Designer and CHOPCHOP design guides by aligning PAM sites only to the reference genome, ignoring any target sites created or altered by genetic variations. CRISPick and CRISPRon use machine-learning methods to improve guide selection, but their predictions rely on reference-sequence properties and do not account for how variations or haplotypes affect on-target efficiency.
CRISPOR goes a step further by annotating guides that overlap known SNPs, but it still selects guides based on the reference genome and does not integrate multiple variations, leaving combined haplotype effects unaccounted for. As a result, genetic variations that affect the guide or PAM sequences can alter or even prevent CRISPR cutting at target locations. This limitation has clinical effects, as evidenced by therapy such as Casgevy for sickle cell disease, where treatment efficacy varies across genetically different groups.
Variant-Aware CRISPR Targeting
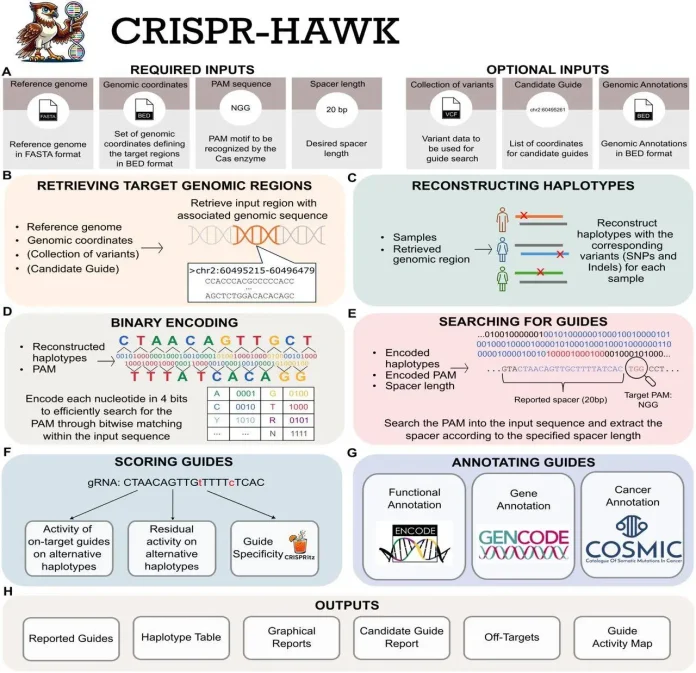
CRISPR-HAWK is a command-line tool that designs, scores, and annotates CRISPR guide RNAs while accounting for real human genetic variation. It employs massive population databases to rebuild individual genomes, so guides are chosen based on a person’s real DNA rather than just the reference genome. The tool looks for guides in user-defined genomic regions, predicts how well each one will work, checks for off-target effects, and includes genetic and functional annotations.
Designing CRISPR Guides with Genetic Diversity
To account for real human genetic diversity, the researchers employed the GRCh38 human reference genome, as well as three important population genetic databases: the 1000 Genomes Project, the Human Genome Diversity Project (HGDP), and gnomAD. Using these datasets, the researchers investigated CRISPR-HAWK on the BCL11A +58 erythroid enhancer, focusing on the clinically improved SpCas9 guide sg1617, which is also used in the approved CRISPR therapy Casgevy.
Input Preparation: Users specify the reference genome (FASTA), target regions (BED), PAM motif, and spacer length. Variant datasets (VCF) and functional annotations are also optional inputs.
Target extraction and haplotype reconstruction: Genomic areas are extracted and extended by 100 kb. Variants are used to rebuild haplotypes; phased variants produce distinct maternal and paternal sequences, whereas unphased variations employ IUPAC codes and combinatorial enumeration. Equivalent haplotypes are deduplicated.
Guide Identification: Haplotype sequences are binary-encoded for faster scanning. Candidate gRNAs are retrieved at PAM sites, with variant-affected bases identified.
Scoring and annotation: Guides are scored for on-target efficiency, residual activity across variants, and specificity using models such as Azimuth, Rule Set 3, Cutting frequency determination (CFD), and Elevation. Functional and sequence features are annotated.
Report generation: It includes tables of guides, variant-aware haplotypes, off-target forecasts, and graphical summaries.
The study discovered that while the sg1617 guide should still work effectively in most people, it may lose cutting ability completely in some people who carry specific genetic variations. This means that a CRISPR therapy based solely on the reference human genome may not work for everyone. Extending the research to other therapeutic and benchmark guides found that 82.5% of reference-designed guides were impacted by at least one variable predicted to alter activity. Overall, CRISPR-HAWK aids in the detection of these conditions, allowing guides to be revised or tailored, resulting in safer and more reliable population-aware genome editing.
Limitations
- As the tool relies on computer predictions rather than actual experiments, the results may not accurately reflect what occurs in real patients. Clinical trials are still necessary.
- The scoring models are more effective for specific CRISPR enzymes (SpCas9 and Cas12a). They may be inaccurate for other CRISPR types or engineered versions.
- It primarily analyzes single-letter DNA alterations (SNVs) and does not completely support larger or more complicated mutations such as indels.
- It does not take into account chromatin structure or epigenetic factors, which can have a significant impact on how well CRISPR cuts DNA in actual cells.
- Genetic databases are not perfect; thus, rare or population-specific variants may be absent, suggesting that some real-world variation may still be ignored.
- It does not predict delivery efficiency, despite the fact that CRISPR’s ability to enter cells can influence editing success.
Conclusion
CRISPR-HAWK demonstrated that genetic variation can influence CRISPR guide performance, particularly in therapeutics. The tool detects when genetic variants may reduce cutting efficiency or alter guide activity by designing and evaluating gRNAs in a haplotype- and variant-aware manner, as shown with the clinically used sg1617 guide in the BCL11A enhancer. Because many reference-designed guides are influenced by real-world variations, population-aware design is critical for reliable and secure genome editing. Although CRISPR-HAWK is based on computational prediction that still requires experimental validation, it represents a significant step toward precise, patient-specific CRISPR therapies.
Article Source: Reference Paper | Availability: GitHub and GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Jainab Shaikh is a postgraduate in Biotechnology with a strong interest in understanding how research translates into real-world innovation. Her areas of focus include biosensors, bioinformatics, and sustainable biotechnological applications. She is passionate about exploring recent scientific advancements and communicating them through clear, engaging, and accessible content. Her work particularly emphasizes research-driven narratives in healthcare, biotechnology, skincare science, and emerging life science innovations.