The discovery of ProtT3 marks a significant recent advance in the fast-developing science of bioinformatics. ProtT3, developed by researchers at the National University of Singapore, the University of Science and Technology of China, and Hokkaido University, combines Language Models (LM) with protein sequence analysis. It provides unparalleled insights into the complicated world of proteins by combining the power of LM with modern protein sequence analysis tools. This new technique has the potential to transform the way scientists and researchers view and analyze proteins, opening the path for substantial advances in bioinformatics and beyond.
Why Protein-to-Text Generation Matters
Proteins are the major macromolecules and are involved in almost all biological activities in the human body, as well as in other forms of life. Still, understanding the sequences and structures of the inherent nucleic acid has remained the biggest challenge. Some of the traditional methods used for the analysis of proteins can be very lengthy, involving detailed and rigorous procedures besides the interpretation calling for professionalism.
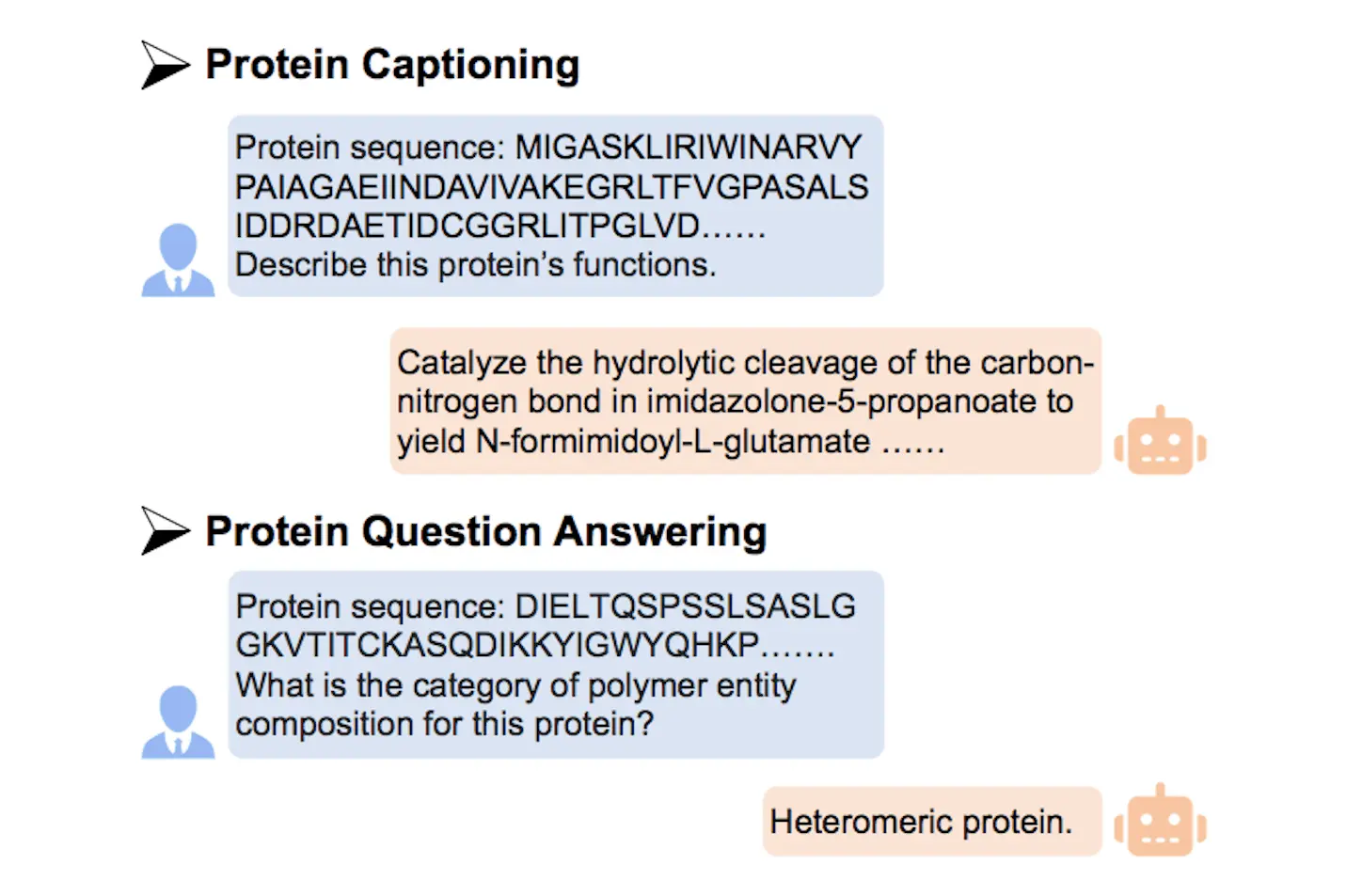
To overcome these challenges, ProtT3 utilizes protein-to-text conversion that can help researchers from different scientific backgrounds utilize protein information. It is a combination of an LM with a Protein Language Model (PLM), and the two modalities are connected using a cross-modal projector that maps the protein embedding into the text space of the LM. This design helps in fending off the LM to function efficiently and work well in protein input data, enhancing performance within protein-based task areas such as protein captioning, protein QA, and protein-text retrieval. This makes access to protein information more open and more collaborative, and it fast-tracks research in fields such as medicine, biotechnology, and pharmacology.
Addressing the Research Gap
Upon reviewing previous studies, Researchers identified two key research gaps:
- Lack of Exploration for Protein-to-Text Generation: Protein-to-texts are inherently a conditional generation task, which means that the LM has to learn to take proteins as the condition for generation. Following that, previous work on cross-modal contrastive learning does not incorporate proteins as primary inputs to the LM directly, and only two approaches, namely, Galactica and ProteinChat, exist in this context. In this case, Galactica employs a very small set of protein sequences to guide its pre-trained models, making it questionable whether it can capture a variety of protein features and functions. Using a linear projector, ProteinChat tries to link protein representations to the text space, but apparently, a more sophisticated approach is needed due to the intricate connections between proteins and texts.
- Missing Quantitative Evaluation: Lack of benchmark often makes it challenging to track progress in protein-text modeling.
Key Contributions
Introduction of ProtT3: First, this study introduced ProtT3, a new machine-learning framework aimed at connecting textual data to proteins. ProtT3 retains a cross-modal projector whereby it combines a PLM for the understanding of proteins and an LM for text processing, hence facilitating translation from protein to text.
Establishment of Benchmarks: Thus, the study provides quantitative standards and discusses potential guidelines and directions for the development of new tasks based on the model of protein description, such as protein captioning, protein QA, and protein-text retrieval. Researchers can access datasets, evaluation scripts, and models pre-trained by us in the experiments described above.
State-of-the-Art Performance: ProtT3 performs highly in all sorts of tasks and makes it possible to ascertain that it is one of the best resources available today. In the case of protein captioning, a BLEU-2 score that is more than 10 points higher than the baseline was obtained when using the Swiss-Prot and ProteinKG25 collections. For the improvement of protein-text retrieval, ProtT3 has demonstrated retrieval accuracy by over 14% on the same datasets. Moreover, it improves the knowledge accuracy for protein QA in terms of the exact match by 2.5% using the PDB-QA dataset.
Model Architecture
It comprises three main components: namely, a Protein Language Model (PLM), a Language Model (LM), and a cross-modal projector.
- Protein Language Model (PLM): Aims at using the ESM-2 model to handle protein sequences, from which it is evident that it is useful in tasks such as protein folding and property prediction. The weights of ESM-2 in the training process are frozen to improve its efficiency.
- Language Model (LM): Using stated Galactica, a decoder-only transformer LM pretrained on scientific papers, they demonstrate a superordinate grounded understanding of the protein concepts.
- Cross-Modal Projector: This allows for bridging the gap between the PLM and LM based on Q-Former, which ensures that the LM readily processes protein inputs by converting the representations into text space.
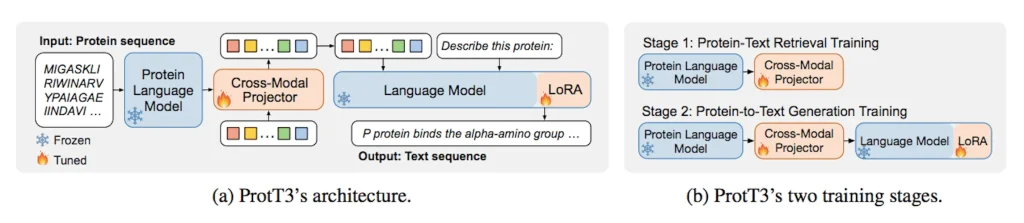
Two-Step Training Approach for ProtT3
The training method for ProtT3 is a two-step training approach devised to improve the abilities of the model in the processing of the protein components and processing text.
- The first stage is known as Protein-Text Retrieval Training, where the model is pre-trained using three objectives (protein-text contrasting, protein-text matching, and protein captioning) inspired by BLIP. These objectives are, however, geared towards the Q-Former’s architecture, which aims to train it to learn relevant features regarding the proteins in the description given from the text data. This stage refines the cross-modal projector’s retrieval process and prepares the projector for the following stage, which is discussed in this section.
- The second stage is the Protein-to-Text Generation Training, which mainly aims to train the model to establish the correspondence between the protein inputs and the text representations. Completing this stage entails transforming protein features into the text space by use of the Language Model (LM) like the Galactica. Incorporating protein understanding into text-based tasks will help the model improve in interpreting and analyzing biological data by incorporating the knowledge into the model.
ProtT3’s Significance and Future Directions
This study represents a new modeling approach for protein-text called ProtT3. It seeks to enable a computationally mediated understanding of proteins through T2P and P2T. To achieve this, ProtT3 incorporates an additional feature, a PLM, into the current structure of an LM for the improvement of the understanding of proteins. They are integrated closely with the help of a cross-modal projector in order to overcome the dissimilarities between the two modules, which belong to different modalities.
For future directions, the researchers propose ablation studies and new research tasks, such as protein captioning, protein QA, and protein-text retrieval; the new ProtT3 model achieves considerable improvements over prior benchmarks. In future work, they propose to advance the ability of LMs to learn 3D protein structures and use them to better disentangle molecular properties, generate new molecules, and improve OOD generalization in drug discovery.
Limitations
- 1D Protein Sequences: This is the case because available proteins greatly outnumber 3D structures and also because the comparison of 1D sequences of amino acids is much simpler. This is because the UniProt database currently offers one-dimensional sequences for 227 million proteins, while the Protein Data Bank has 220,000 three-dimensional structures. This, in turn, enables us to sample a greater volume, assembling 1 million protein-text pairs, which is further than the 143,000 pairs utilized in previous analyses deploying 3D structures. Protein folding takes four months to predict the 3D structure to about 440,000 sequences on an A100 80GB GPU; thus, dealing with only 1D sequences and 3D modeling is reserved for future studies.
- Dynamic Protein Structures: Proteins possess versatile structures that cannot be described accurately by natural language. However, like the work done in video-to-text generation, where language partially describes dynamic videos, the work done in this paper on protein-to-text generation is a step in the right direction of translating a wide array of biological occurrences into an understandable form.
Conclusion
ProtT3 can be considered as the further evolution of language models for proteins based on sequence analysis. When combined with PLM integration and cross-modal projection, this approach opens the door to new LMs and new ways of thinking about and analyzing biological data. Continuing the trend of presenting exciting connections between AI and bioinformatics, ProtT3 is a shining example of the applied value of cross-disciplinary study and development.
To learn more about ProtT3, kindly refer to the Reference Paper. Its code is available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.