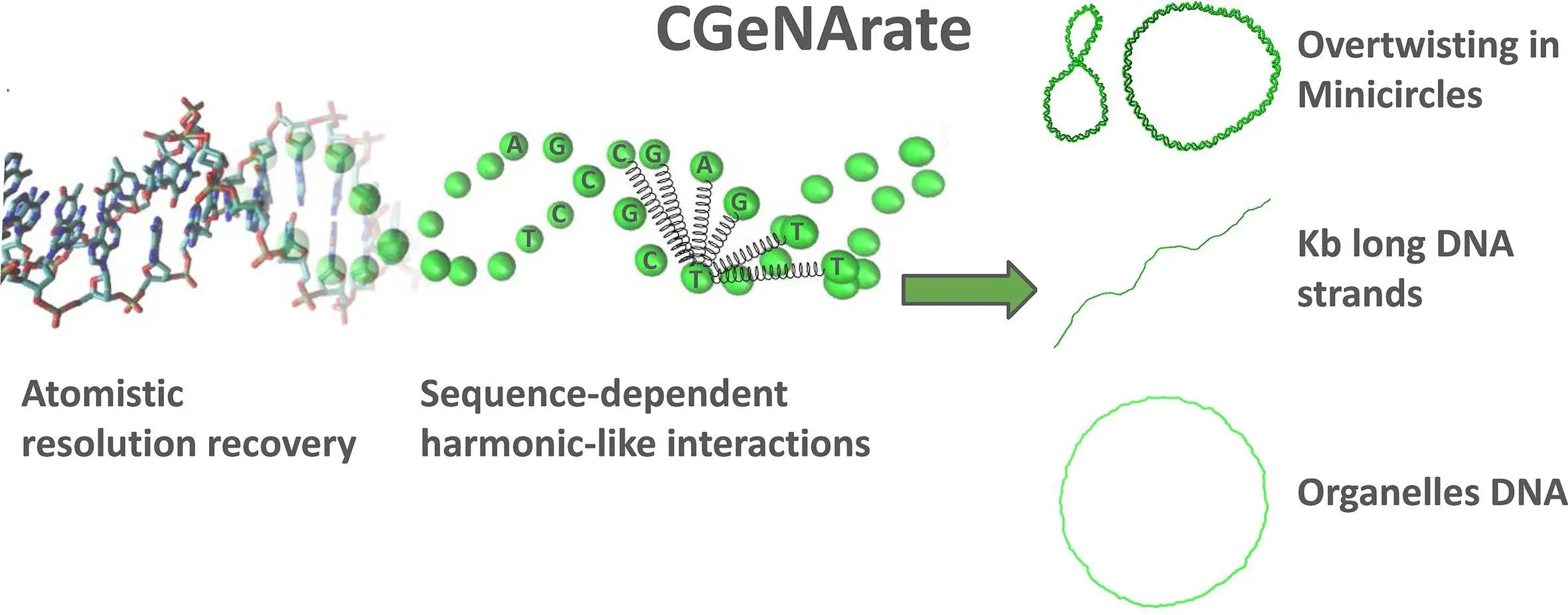
Researchers from the Institute for Research in Biomedicine (IRB Barcelona) introduce CGeNArate, a short sequence-dependent coarse-grained model for DNA simulations. CGeNArate solves the challenge of simulating long duplexes of DNA (kilobase-long) accurately without using a lot of computer power in the process. The model accurately reproduces both local and global DNA properties, making it a valuable tool for biotechnology and chromatin research. This advancement promises to accelerate studies in genetics and cellular biology by enabling more efficient and comprehensive DNA simulations.
DNA is the molecule that holds the instructions for life. It’s a fascinating process of complex structures and interactions. Nevertheless, its dynamics, especially over long dimensions, require a lot of computational time. CGeNArate enables rapid and precise DNA simulations, facilitating detailed observation of genomic structural modifications.
Turning on the Light on Difficulties in DNA Simulations
Think of DNA as a long, intricate bead string. A single bead represents each unit within the structure of the DNA whose interactions and movement we must comprehend to understand how it works. In traditional terms, scientists employ atomistic simulations to model every atom in the DNA molecule. While these simulations are highly detailed, they become very expensive in terms of computer time when one deals with large stretches of DNA. In this light, our ability to study gene regulation and other processes involving extensive segments of DNA is impeded.
Enter CGeNArate: A Simpler Yet Powerful Approach
CGeNArate offers a smart alternative. It uses a coarse-grained method where fewer beads represent DNA as compared to atomistic simulations. By doing so, this simplification allows for faster computations while maintaining valuable data about the behavior of DNA. However! The CGeNArate is not just an additional simplified model that it seems to be. It utilizes the potential possessed by machine learning (ML) to close the gap between the coarse-grained model and the detailed atomistic world.
Here is how it works:
Coarse-Grained Representation: CGeNArate lumps together several atoms in the DNA molecule into a single bead. This reduces the count of interacting units and speeds up simulations.
Sequence-Dependence: Unlike some coarse-grained models, CGeNArate considers the specific sequence of DNA bases (A, C, G, T). This ensures that each model can capture the unique properties of different regions of DNA.
Machine Learning Magic: After the coarse-grained simulation, CGeNArate employs machine learning to translate the coarse-grained data back into a detailed, atomistic representation. The retrieved atomistic trajectory is very similar to what would be produced using traditional all-atom simulations.
Unveiling the Power within CGeNArate
CGeNArate’s magic lies in its amazing ability to accomplish two opposing objectives:
Speed: Compared with traditional atomistic methods, simulations run much faster, enabling researchers to analyze more sections of DNA in less period.
Accuracy: The ML-powered reconstruction produces atomistic details with high accuracy, making them comparable to those obtained from high-quality all-atom simulations.
This efficiency-accuracy combination opens the door to new possibilities:
Large-Scale Chromatin Dynamics Study: Chromatin is a tightly packed form of DNA within the nucleus that is important in gene regulation. CGeANArate allows researchers to model entire genes and their interactions within the chromatin environment, providing unprecedented insights into gene control mechanisms.
Modeling DNA-Protein Interactions: Proteins are important in controlling DNA function. CGeNArate’s simulation of bigger chunks of DNA makes it possible to study proteins’ interaction with and their effect on DNA dynamics at a higher level.
Revealing the Secrets Behind Long-Range Effects: Even when physically distant, DNA sequences can influence each other. By simulating larger stretches, CGeNArate makes it possible to explore these long-range interactions in DNA.
Conclusion: A New Dawn for DNA Simulations
The CGeNArate project is an example of how creative modeling technologies mixed with machine learning can create powerful tools for the whole of science. By providing a faster and more accurate way to simulate very long DNAs, it introduces us to a new era of genomics research. The possibilities that CGeNArate opens are limitless, from decoding gene regulation and protein-DNA interactions; this implies its use in various fields by different scholars.
This innovation should not be seen as the ultimate result but instead as a transition to other possibilities. We are going through significant advancements in computation power, together with a better comprehension of DNA dynamics, which will lead to the development of more advanced coarse-grained models. The future of bioinformatics is bright, with CGeNArate showing us the way forward toward a deeper understanding of the wonderful world that is our DNA.
Join the Conversation
The development of CGeNArate is an important step forward for DNA simulations. CGenArate has the potential to fundamentally change our understanding of various biological processes due to its capability to analyze large-scale dynamics of DNA with great precision and speed.
Article Source: Reference Paper | Reference Article | The executable for CGeNArate is also available in the public repository https://mmb.irbbarcelona.org/gitlab/dfarre/cgenarate-materials.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.