The discovery of novel molecules with desired properties is a long-standing challenge in medicinal chemistry. On the other hand, de novo drug design tools have been developed as a result of recent developments in machine learning. In spite of the increasing popularity of these technologies, user-friendly and adaptable resources are not available. Scientists from Leiden University in the Netherlands present DrugEx, a new versatile open-source software package for multiobjective reinforcement learning in de novo drug design. DrugEx combines various generator architectures, scoring tools, and optimization methods, providing researchers with a flexible and customizable platform.
The Challenge of Drug Discovery
Drug discovery involves intricate and resource-demanding procedures that can span over several years and incur significant financial expenses. Computer-aided drug design helps expedite this process by identifying promising compounds with higher success probabilities. De novo drug design (DNDD) involves exploring the vast chemical space to discover hit, lead, and future drug candidates. With approximately 1063 molecules in the drug-like chemical space, DNDD presents a promising approach to discovering new therapeutic compounds.
The Influence of Machine Learning
The field of DNDD has been greatly influenced by rapid technological improvements and the popularity of advanced machine-learning methods. Modern approaches such as population-based metaheuristics, recurrent neural networks (RNNs), generative adversarial networks, variational autoencoders, and transformers have transformed DNDD. Transfer learning, conditional learning, and reinforcement learning (RL) are also applied to generate molecules with desired properties.
DNDD inherently involves multiobjective optimization (MOO) due to the diverse set of objectives guiding the drug discovery process. These objectives include maximizing predicted efficiencies, synthetic accessibility, drug-likeness, and minimizing off-target effects and toxicity. DrugEx addresses the MOO challenge and provides a comprehensive framework for optimizing multiple objectives simultaneously.
DrugEx: A Comprehensive Software Package
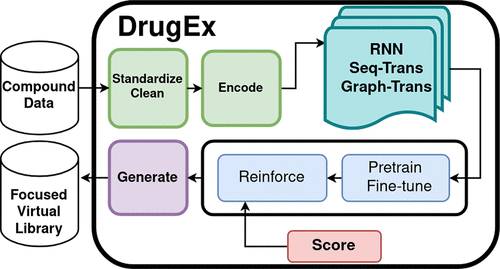
DrugEx is an open-source software package that consolidates and redesigns scripts from previous DrugEx papers. It offers multiple generator architectures, a variety of scoring tools, and various optimization methods. Featuring a flexible application programming interface (API), the package accommodates multiple user preferences, enabling them to engage with it through the command line interface (CLI) or the visually intuitive graphical user interface (GUI) GenUI. DrugEx also provides pre-trained models to facilitate the de novo design of molecules.
DrugEx incorporates four generator architectures: two SMILES-based RNN models using GRU or LSTM units and two fragment-based transformer models using either sequences or graphs as molecular representations. These models enable the generation of molecules based on different input formats and provide options for incorporating stereochemistry and building blocks.
DrugEx offers default data preprocessing steps, including standardization and fragmentation, using algorithms like BRICS or RECAP. The package supports pretraining and transfer learning to familiarize the generator with the language of drug-like molecules and guide it toward the desired chemical space. Training involves assessing loss on a separate test set, allowing for early stopping and fine-tuning of the generator.
Scoring compounds in DrugEx involves three stages:
- Obtaining raw scores for each objective
- Scaling the scores with modifier functions
- Performing multiobjective optimization
DrugEx provides a range of predefined objective functions, including QSAR/QSPR models. Modifier functions transform objectives into maximization tasks and scale scores between 0 and 1. Multiobjective optimization can be achieved through aggregation or Pareto ranking-based schemes.
Conclusion
DrugEx is an open-source software package that enables the training of diverse generative models for the de novo design of small molecules. The researchers look forward to this package as a significant advancement in the area and will aid in the development of better AI-powered models and tools. Future developments will focus on integrating new objectives and alternative compound representations, as well as improving user-centric features and GenUI integration. The authors hope DrugEx will help overcome challenges in integrating AI tools into drug discovery workflows.
Article Source: Reference Paper | DrugEx: Package is publicly available at GitHub
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.