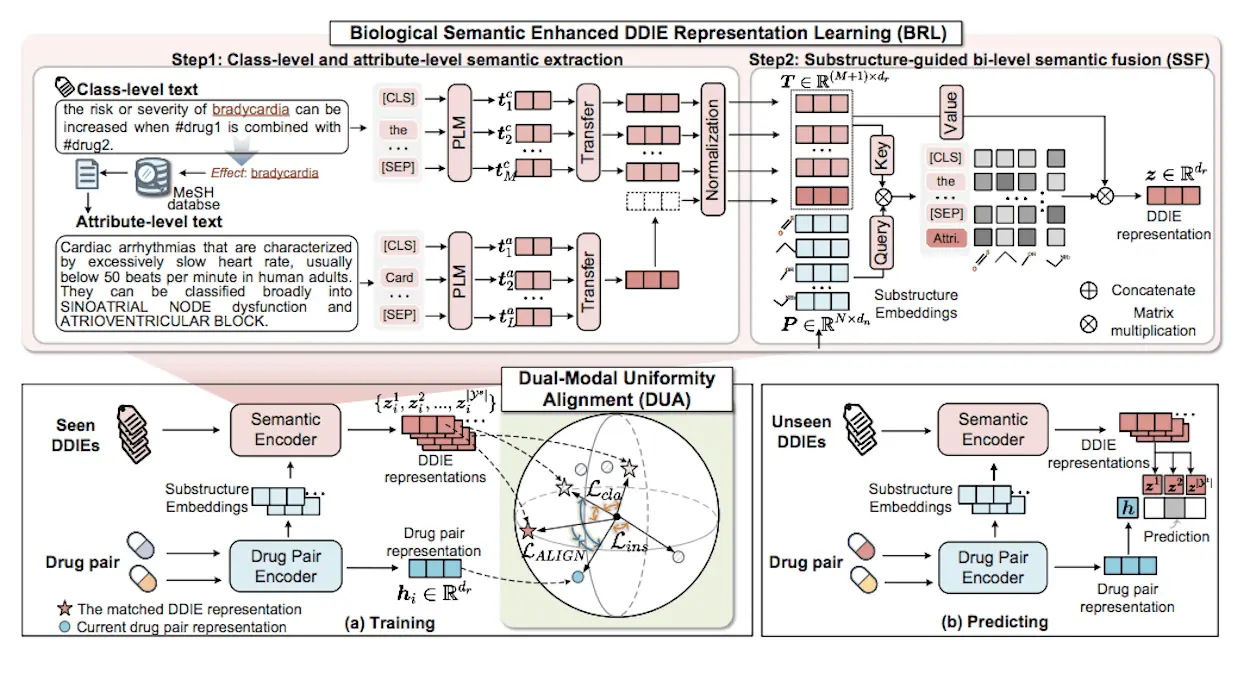
In the context of the field of medical practice, identifying potential DDIs (Drug-drug interactions) is critical in assessing the safety of a patient’s prescribed medications and their precise outcomes. Some of the DDI events (DDIEs) are not directly observable, making it cumbersome to use the traditional methods; hence, researchers from the College of Informatics, Huazhong Agricultural University, Wuhan, called for better ways and introduced ZeroDDI, a novel method designed for the zero-shot DDIE prediction task, where unseen DDIEs with no labeled instances in the training stage are classified.
The Challenge of Drug-Drug Interaction Prediction
The identification of DDIs has remained among the most complex tasks in the healthcare setting due to the various interactions that are involved when different drugs are used. The large number of potential DDIEs belonging to different classes also contributes to it; therefore, it is crucial to accurately classify DDIs to detect possible changes in pharmacology. With most DDIEs buried beyond the surface, traditional techniques prove ineffective and have prompted the emergence of new techniques like Zero-Shot Learning to boost the prediction of drug interactions, given the dynamic nature of the drug fields. Scientists conduct their analysis to find more reliable approaches and strategies that will help predict DDIs and consequently resolve the problems associated with polypharmacy.
Zero-Shot Learning: A New Paradigm
Zero-Shot Learning (ZSL) is a revolutionary concept in machine learning, especially in vision and healthcare domains. Thus, applying ZSL methods, all instances from the unseen classes can be classified since all the classes are in the same semantic space. It makes it possible to predict other new cases that are not included in the training data to perform new classifications that were not known before, transforming classical problems of classification. In the context of healthcare, ZSL can be useful for improving outcomes resulting from drug-drug interaction prediction along with other complex issues in the healthcare domain due to the usage of class attributes, names, and textual descriptions to enhance the prediction. The use of ZSL shows that we have entered a new generation of machine learning, in which models are capable of improving and performing on different problems, opening the door to AI’s improvements.
Semantic Enhanced Learning
Semantic enhanced learning is defined as the process of incorporating semantic knowledge into ML approaches to improve the former’s comprehension as well as capability on a particular task. By using semantic knowledge, such as class-level and attribute-level semantics, the formation and characteristics of the semantics of the model can be captured more effectively. It improves the discriminative capability of the models as well as allows them to transform more salient and semantically related features for applications such as drug-drug interaction prediction. Semantic enhancement of learning is also useful in the higher degree of the specialization of the machine learning systems as well as the level of interpretability within the given area of application.
Dual-Modal Uniform Alignment
DUA is another compatibility function developed to look for drug pair similarity and the contracted DDIE representation to classify the drug pairs. This alignment function ensures that both the structures, drug pairs and the corresponding DDIEs in the text modal lie uniformly distributed on the unit sphere. In this way, DUA resolves the problem of blurred decision boundaries resulting from class imbalance and optimizes the performance of zero-shot drug-drug interaction prediction models. DUA strategy improves the interpretability of the models and their discriminative capacities, hence increasing the ability of the models to solve complicated classification problems in healthcare and other fields.
Methodology
The methodology used focuses on the training data S that contains drug pairs and their matched DDIE label; the set of seen and unseen DDIE labels is defined as Ys and Yu, respectively. Testing data T contains drug pairs and test DDIE labels belonging to unseen classes as well as both seen and unseen classes. Evaluation methods include Common Zero-Shot Learning (CZSL) and Generalized Zero-Shot Learning (GZSL) paradigms, and in terms of evaluation measures, the work used top-k accuracy. Hyper-parameter sensitivity analysis also looks at the effects of temperature and weight loss parameters such that users must opt for the best parameters to gain the best results. The study is focused on the comparison of class-based and attribute-based models for cost analysis and underlining the importance of the approach for Zero-Shot Drug-Drug Interaction Event (ZS-DDIE) cases.
Comparison of Results and Evaluation of Efficiency
With the help of ZSL hence, a method called ZeroDDI has outperformed other metrics in the experiments designated as Conventional Zero-Shot Learning (CZSL) and Generalized Zero-Shot Learning (GZSL). The bi-level fusion applied selectively and the semantic fusion based on the molecular substructures have also been seen to improve the performance of the DDIE prediction models.
Advantages and Applications
In general, the approach of semantically enhanced learning and dual-modal uniform alignment has the following advantages in the field of DDI prediction and other similar tasks. Incorporating the semantic information enhances the discriminative nature of the models and their performance in the case of map accuracy of novel DDIEs. Such advances help not only to improve the patient’s safety by identifying their possible adverse interactions but also to progress the concept of individualized medicine and pharmacovigilance.
It is worth mentioning that the application of these methodologies does not limit itself to fields of healthcare only; however, it applies to all domains that involve classification and prediction expertise. Application areas like finance, e-commerce, and cybersecurity are a few areas where you want your machine learning models to be better than before by using more semantic information and alignment techniques. Due to the flexibility of the given methods and their high efficiency in solving various classification problems, these methods can be considered effective tools and contribute to the development of the core technologies of machine learning in multiple disciplines.
Conclusion
The field of drug-drug interaction prediction is headed towards improved models with the help of Zero-Shot Learning techniques and representations enhanced with bio-information. These solutions can potentially improve the quality of care offered to patients and the safety of drugs in the health field.
Article Source: Reference Paper | ZeroDDI code is available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.