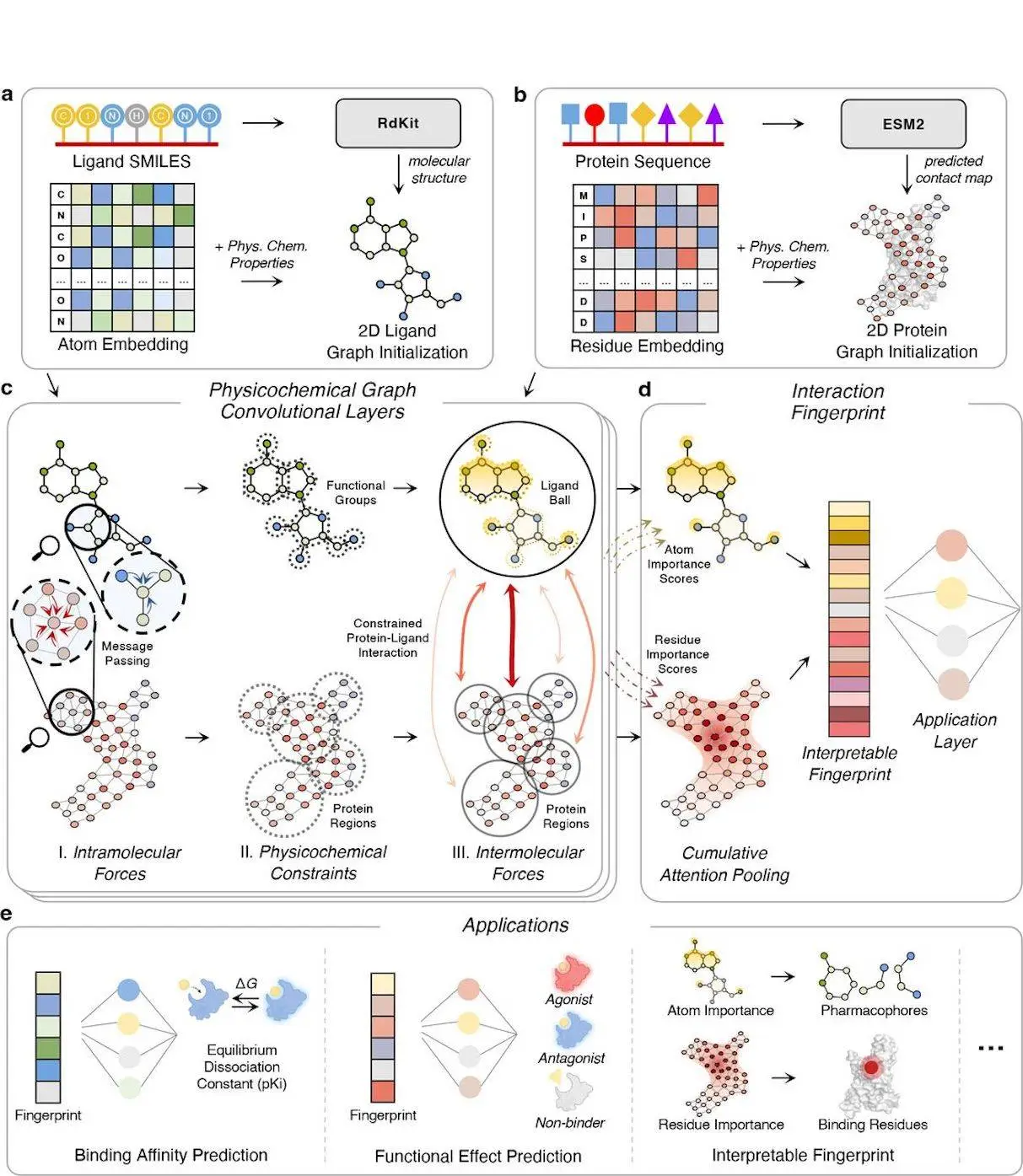
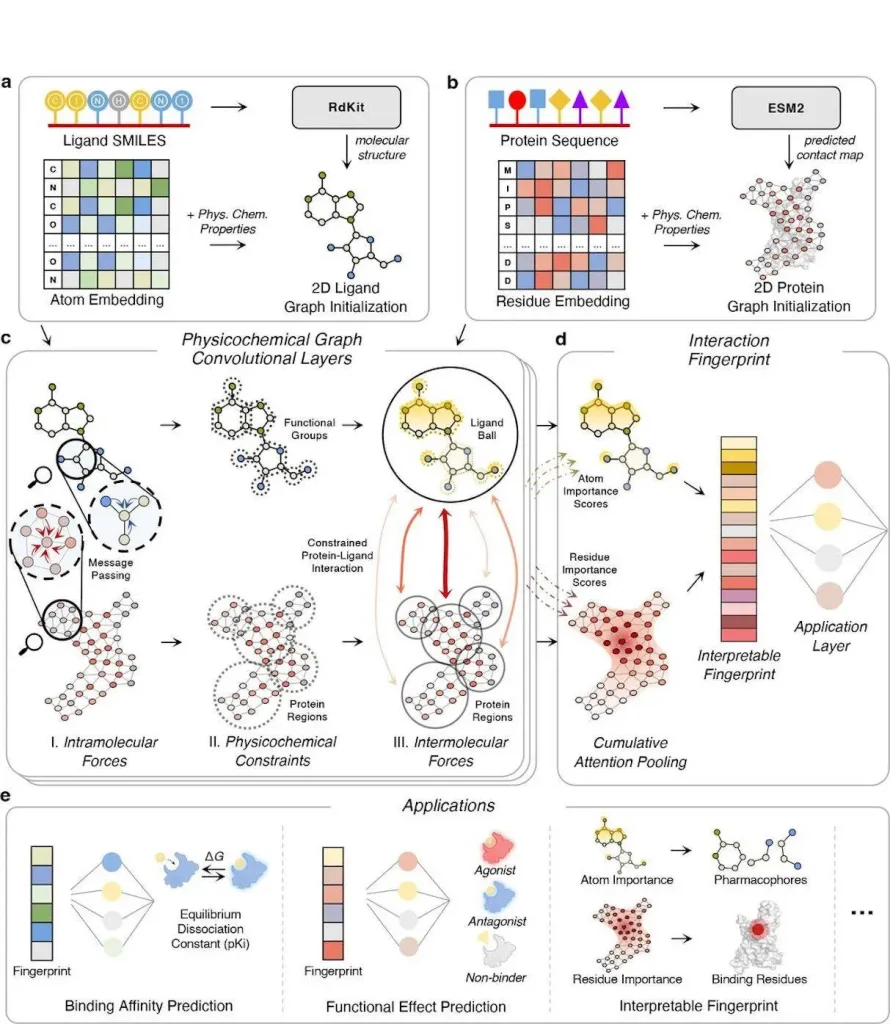
Improved identification of interactions between proteins and ligands has been invaluable in the progress of computational biology, potentially enhancing drug discovery and revealing more about the molecular world. Traditional approaches tend to utilize structural information, which is sometimes scarce or nonexistent. Enter PSICHIC (PhySIcoCHemICal): a graph convolutional network model, physicochemical and chemotherapeutic interactions in sequence learning of protein-ligand interactions. This pioneering approach offers the potential to revolutionize our outlook to predict the interactions in the application context as envisaged in research and industry.
PSICHIC Architecture
PSICHIC is a complex model which can be used to anticipate protein-ligand interactions based on the input of sequence information. To do this, it begins ligand and protein graphs from corresponding SMILES strings and protein sequences, including physicochemical properties. PSICHIC then performs the integration of the multiple forces through the use of GNNs where the inter and intramolecular forces are entangled through physicochemical graph convolutional layers with constraints and MinCUT loss. Thus, in three learned steps of intermolecular force modeling together with attentional aggregation and cross-attention, PSICHIC creates an interaction fingerprint easily interpretable by the user, which predicts and interprets potentially rich interaction properties with higher accuracy and insight, significantly enhancing drug discovery and virtual screening.
PSICHIC Optimization
PSICHIC is optimized using the Adam optimizer with some specific parameters set to be β1=0.9 β2 = 0.999, ε = 10−8, and the Static learning rate is 10−4. The possible values of the gradient in the global version are bounded by 1.0. The decay is set to 0, and weight decay is set to 10−4 for regularization. In large-sized datasets such as PDBBind v2016 and other large-scale interaction data, optimization is fine-tuned to address the problems that may arise due to size and distribution to enhance stability in learning. A virtual machine is used for the training process, where the host machine contains an NVIDIA A100 Tensor Core GPU for high computation and speed.
Pharmacological Materials
The main pharmacological materials used in the study were thus CHO Flp-INTM cells, DMEM, DMEM high glucose medium, FBS, Hygromycin B, and ADA, which were sourced from the following suppliers: These materials were crucial as a precursor to pharmacologically act out the concept in the new approach of A1R agonists and the data analysis of the research. The study also gave credit to many organizations and facilities for the comprehensive screen and computational needs.
PSICHIC Framework
The PSICHIC is a highly elaborate model that is meant to predict protein-ligand interactions through the use of sequence information. It includes Physicochemical properties and employs physicochemical graph convolutional layers to address forces of intramolecular and intermolecular. In particular, using graph neural networks (GNNs) and attention, PSICHIC synthesizes interpretable interaction fingerprints, which effectively capture and predict interaction properties. This framework completely transforms the drug discovery process as it furnishes pharmacologists with the means to identify the phenomenon of molecular interactions with enhanced accuracy and understanding of the underlying, thereby significantly improving the field of pharmacology.
Effectiveness of Interaction Fingerprints
The interaction fingerprints generated from PSICHIC have revealed them to be very efficient in predicting and analyzing the protein-ligand interaction. These can be beneficial in describing binding affinity and functional effects using the sequence data exclusively, which helps integrate fingerprints and other computational tools. Such an approach evidentially works well in unraveling the physicochemical interaction mechanisms and optimizing them by identifying the essential ligand functional groups by the PSICHIC tool. In conclusion, the inherent interpretability and efficiency of these interaction fingerprints render PSICHIC significant for drug discovery and virtual compound library screening.
Generalizability of Interaction Fingerprints
The approach, based on applying interaction fingerprints, makes PSICHIC suitable for the screening procedure completed for the identification of potential drugs, including A1R agonists that play a vital role in cardiovascular and neuronal disorders. Through testing on multiple libraries of compounds, PSICHIC highlights its capability of providing predictions of and insights into molecular binding behavior, facilitating the identification of effective therapies. The designed model can effectively decode isolated physicochemical mechanisms and accurately predict the inferring binding affinity labels, especially for large chemical space, without prior knowledge of the binding sites; thus, it is particularly useful for targeted in silico screening and drug discovery.
Conclusion
PSICHIC is located right on the boundary of the new generation of tools in computational biology, where graph neural networks and physicochemical properties are powering the new opportunities for protein-ligand interaction prediction based on the sequence data. Its prospects for drug development and biological research are primed to impact the speed of science creation and human well-being positively. Consequent inventions such as PSICHIC offer a glimpse of how the field will continue to progress and will be critical in advancing bioinformatics.
Article Source: Reference Paper | The source code and data files for retraining and evaluating PSICHIC are available on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.